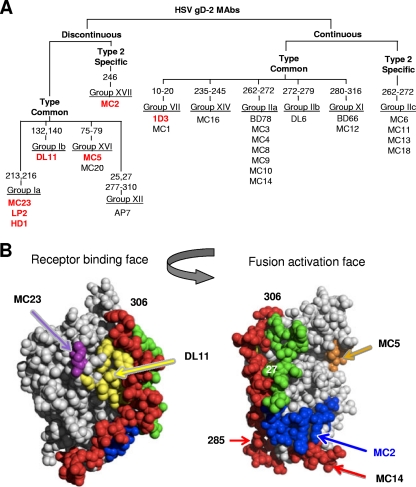
Fig 1.
Grouping of MC series of MAbs to gD and their neutralization properties. (A) Tree-like diagram showing how the MC MAbs relate to each other and to previously characterized anti-gD MAbs (22, 37). MC MAbs were grouped according to several criteria, starting with whether they react with any of a series of overlapping peptides to gD2. Additionally, we used truncated forms of gD to delineate the C-terminal end of the discontinuous epitopes. In some cases, the numbers refer to the amino acid changed in monoclonal antibody-resistant (mar) mutants as well as to the position of linker insertion mutations found to affect MAb binding to gD (11). Western blotting of gD proteins was used to determine MC MAb type specificity. They were compared to monoclonal antibodies that had previously been characterized and grouped using biosensor competition as well as being grouped according to which peptides they recognized. Previous groups (group I to group XI) were maintained, and new groups (XII to XVI) were added to accommodate MAbs that did not fit into known groups (37). Those shown in bold and in red have virus-neutralizing activity. MC12 recognizes the same amino acids as the previously mapped MAb BD66 and was placed in group XI. MC1 recognized synthetic peptide 1-20 and was placed in group VII. Eleven type-common MC MAbs were added to group IIa (previously defined only by BD78). Five new groups (IIc, XIV, XV, XVI, and XVII) accounted for the remaining MC MAbs. (B) Structures of gD showing the receptor-binding face and the fusion activation face.