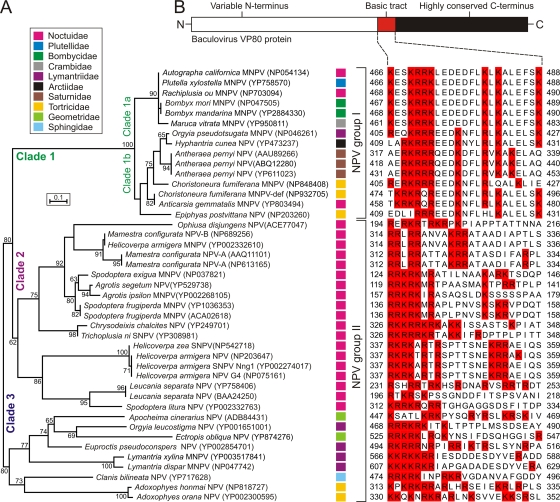
Fig 1.
Overview of VP80 phylogeny and sequence conservation. (A) Unrooted ML-tree built up from VP80 protein sequence data. Forty-four VP80 homologous sequences were aligned with the help of ClustalX. All positions containing gaps and missing data were eliminated from the data set, and phylogenetic analysis was conducted in MEGA4. Numbers at branches denote percent bootstrap values (out of 500 replicates), and numbers in the brackets indicate the GenBank accession number. Color squares at each baculovirus strain indicate host lepidopteran family as explained in legend (top frame). (B) All VP80 proteins contain a tract of basic amino acid residues. Schematic representation of VP80 sequence conservation (top scheme) with a list of aligned VP80 basic tracts. Basic amino acid residues are highlighted in red boxes. The alignment was constructed using ClustalX, manually edited in GeneDoc, and coloring was performed in JalView.