Fig 2.
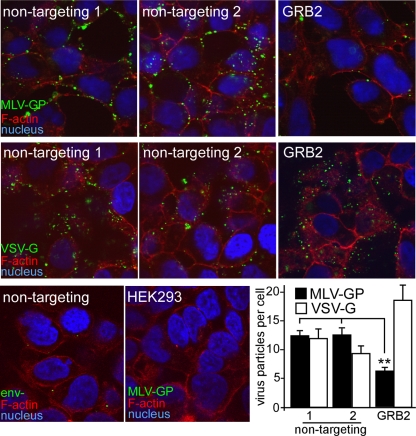
Suppression of endogenous GRB2 inhibits eMLV binding. Fluorescent eMLV particles were generated by producing virus particles containing a fusion of the MLV matrix protein coupled to GFP. Virus particles were purified by pelleting through a 20% sucrose cushion and titrated on HEK293 cells expressing mCAT-1–HA. siRNA against GRB2 or nontargeting siRNA (Ambion, Qiagen) was transfected into cells, and virus binding was measured after 72 h. Cells were first chilled to 14°C to prevent endocytosis, and GFP-tagged eMLV gp85 (top panels)- or VSV-G (middle panels)-pseudotyped particles were added for 1 h. Unbound virus was removed by rinsing with DMEM, and cells were fixed in paraformaldehyde. In order to visualize the cell outline, the nonpermeabilized cells were treated with Alexa Fluor 594-conjugated phalloidin. Under these conditions, some stain permeated through the cell membrane and stained cortical actin. Cell nuclei were stained with DAPI. Samples were then examined by using confocal microscopy. As a control for the nonspecific binding of particles, particles lacking an envelope glycoprotein (Env−) or eMLV g85-pseudotyped particles were incubated with mCAT-1-expressing cells or HEK293 cells lacking a receptor, respectively (bottom panels). The number of virus particles bound to the cells was determined by using CellProfiler image analysis software to count virus particles associated with each cell. The average numbers of viral particles per cell (with standard deviations) are shown and represent results obtained from 10 random images, each containing >20 cells for each individual treatment (bottom right). ∗∗, P < 0.01, determined by one-way ANOVA.