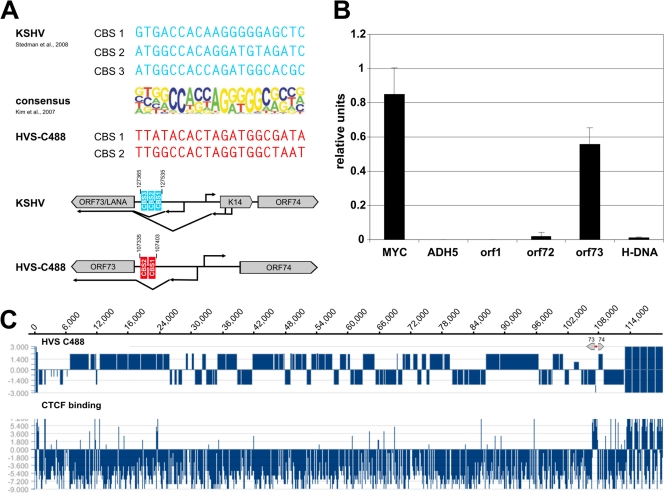
Fig 1.
Detection of putative CTCF binding sites within the intergenic region of HVS orf73/orf74. (A) Schematic comparison of published CBS within the intergenic regions of KSHV orf73/K14 (AJ410493; blue) and potential CBS at HVS orf73/orf74 (KSU756098; red) to the consensus binding sequence of CTCF (23). (B) ChIP analysis of CTCF binding to the intergenic region of HVS orf73/orf74 (exemplary analysis by B. Alberter). Viral genomes from HVS transformed human CBLs were analyzed in three independent ChIP experiments using an anti-CTCF serum (rabbit) and subsequent SYBR green PCR. Mean values were calculated for each of the three data sets, and the mean values of the single experiments were then normalized to form a combined mean value (= 1). CBS of the myc gene locus served as positive control and the first intron of the cellular ADH5 gene represents the negative control. Further coding regions of HVS (orf1 promoter region, orf72 and H-DNA) were analyzed in addition to the putative CBS at the intergenic region of orf73/orf74. (C) ChIP-on-Chip analysis of CTCF binding to the latent HVS-C488 genome using a custom oligonucleotide array (done by B. Alberter as described in reference 2). Upper panel: HVS strain C488 L-DNA containing the known 75 open reading frames and five H-DNA repeat units. The scale of the x axis corresponds to the genome position in base pairs. Lower panel: CTCF binding in the latent HVS genome. The ratio of Cy3-labeled input and Cy5-labeled ChIP probe signals along the genome are displayed using SignalMap version 1.9.