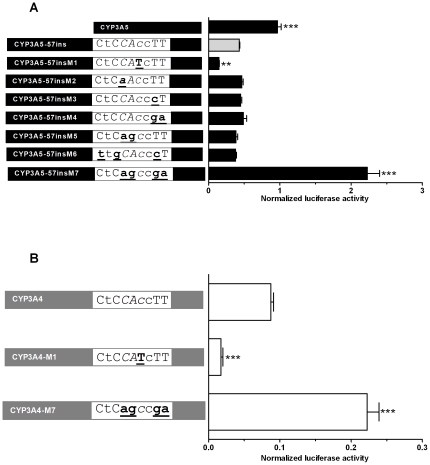
Figure 7. Mutational analysis of the CYP3A4-derived YY1 binding site expressed (A) in MDCK.2 cells in the CYP3A5-57ins promoter construct and (B) in LS174T cells in the native CYP3A4 promoter.
The uppercase and lowercase letters represent preferred and tolerated nucleotides, respectively. The bolded and underlined letters indicate the mutated nucleotides. The mutations either restore the consensus core motif (M1), or progresively disrupt the YY1 binding site (M2 to M7). The construction of CYP3A4 and CYP3A5 mutants is described under “Materials and Methods.” Promoter-driven firefly luciferase activities were normalized using activities of the co-transfected renilla luciferase driven by a constitutive promoter and compared in (A) to that of the CYP3A5-ins57 construct and in (B) to that of the wild-type CYP3A4. Data are expressed as mean values (±SEM) of four to eight independent experiments, conducted as triplicates. Statistically significant differences are indicated by asterisks (** p<0.01,*** p<0.001).