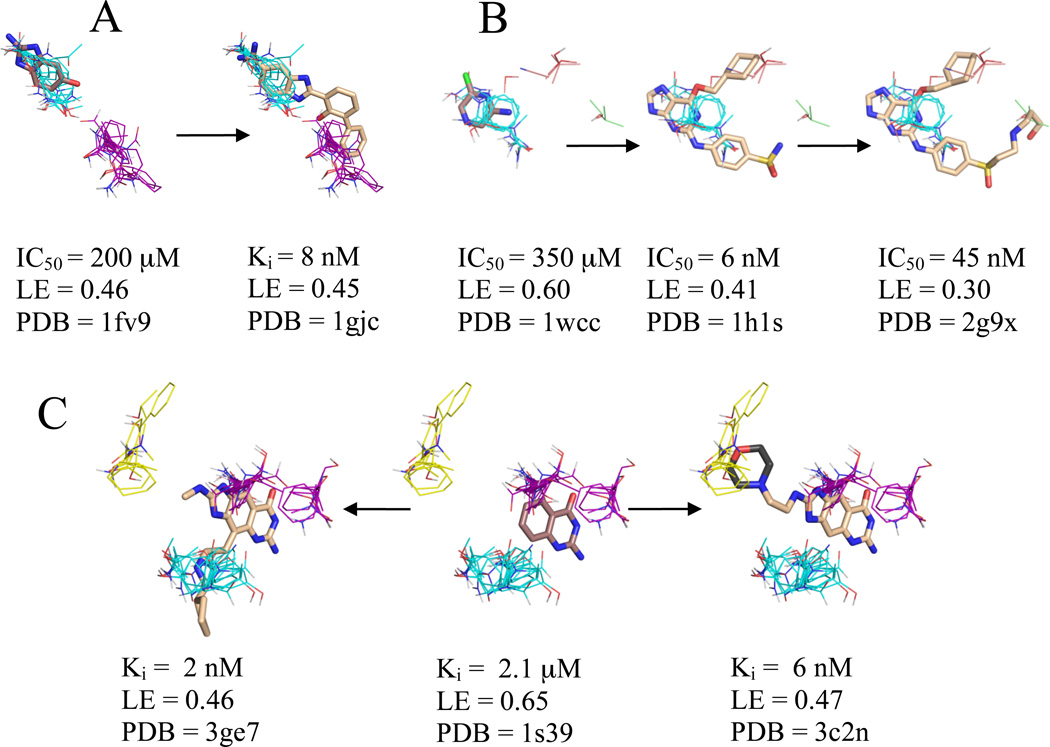
Figure 2.
Significant consensus clusters in the binding site, predicted by computational mapping, for unbound (A) urokinase (2o8t), (B) CDK2 (1hcl), and (C) TGT (1pud), represented as lines with ligands represented as sticks from aligned bound structures. Affinity measures, ligand efficiency (LE), and the PDB code of the bound structure are listed for each ligand. Fragments identified as cores by Congreve et al.9 have carbons colored brown, and higher affinity ligands have carbons colored wheat where electron density supports the position of the atoms and dark grey where the atoms are a representative of a number of possible conformational states. Consensus cluster carbons are colored by rank as follows: 1st - cyan, 2nd - purple, 3rd - yellow, 4th – salmon, and 6th - pale green. TGT’s core fragment is the only example of a core fragment that does not overlap the top ranked consensus clusters due to a substantial change in the conformation of Tyr106.