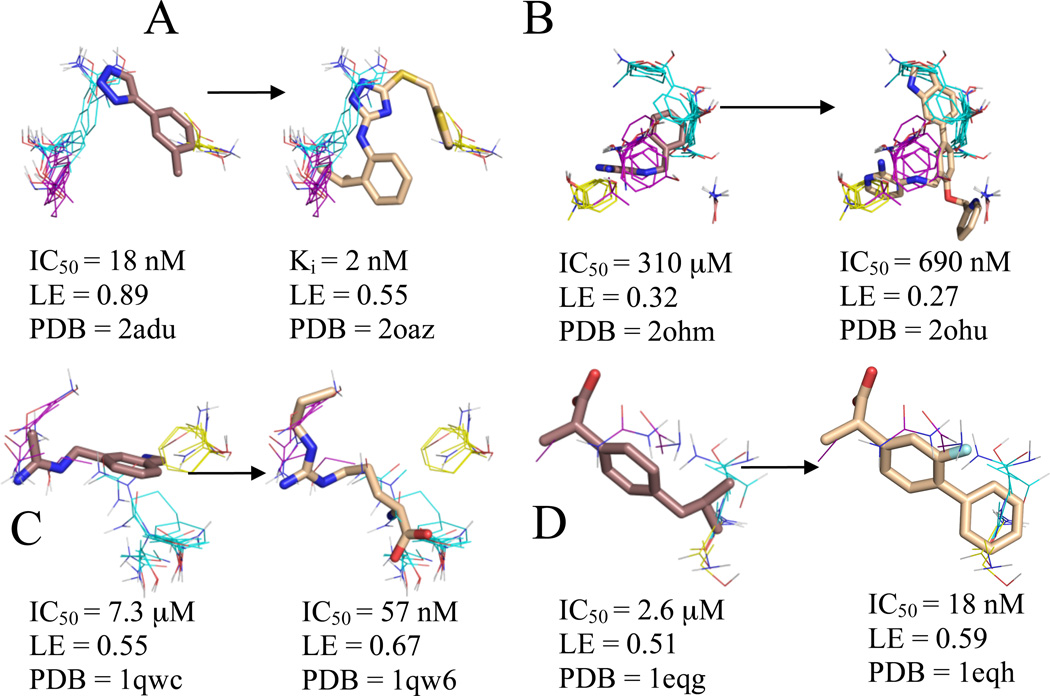
Figure 3.
Significant consensus clusters in the binding site, predicted by computational mapping, for unbound (A) MetAp2 (1bn5), (B) BACE (1w50), (C) nNOS (1zvi), and (D) COX-1 (1prh), represented as lines with ligands represented as sticks from aligned bound structures. Affinity measures, ligand efficiency (LE), and the PDB code of the bound structure are listed for each ligand. Fragments identified as cores by Congreve et al. have carbons colored brown, and higher affinity ligands have carbons colored wheat. Consensus cluster carbons are colored by rank as follows: 1st - cyan, 2nd - purple, and 3rd - yellow. All four core molecules overlap to some extent with the top consensus cluster, and BACE, COX-1, and nNOS have more inhibitor atoms placed within the core consensus clusters than the cores themselves. This is especially noteworthy since COX-1 and nNOS were the only two systems that had an inhibitor with higher LE than the LE of the core moiety and were also the only two systems that lacked extension clusters, and hence the core has been evolved into lead compounds within the main consensus cluster.