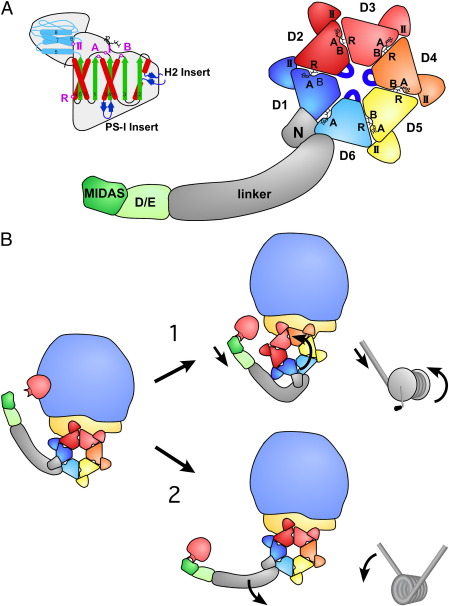
Fig. 4.
Domain organization of Rea1. A, Rea1 consists of an N-terminal domain, six ATPase modules that form the ring domain, followed by a long α-helical linker, a D/E rich region and a MIDAS domain. The predicted structural organization of an ATPase module: the central α/β domain is shown in red (α-helices) and green (β-strands), including Walker A (A), and Walker B (B) motif, sensor-I (I) and the arginine finger (R) indicated by a purple dot. The lid domain including the sensor-II (II) region is depicted in light blue. The clade-specific helix2 insert and pre-sensor-I insert (PSI) are indicated in dark blue (colors are identical to secondary structure prediction of Figs. S5 and S6.) B, Possible mechanisms of Rea1-mediated release reactions. Mechanism 1 displays a ratchet-hoist model, where energy is utilized to move the Rea1 ATPase ring on the pre-ribosome, thus creating a tension that pulls off the substrate. In mechanism 2 (power-stroke model) the Rea1 ring domain stays firmly attached at the pre-ribosome. Conformational changes within the ATPase ring are causing a power stroke: i.e. an active movement of the linker domain with the attached D/E rich region and the MIDAS domain.