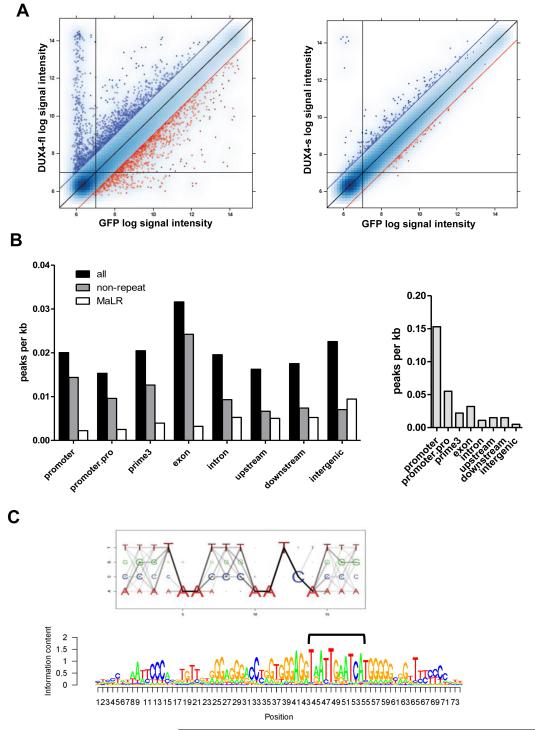
Figure 1. DUX4-fl activates the expression of germline genes and binds a double-homeobox motif.
(A) Pairwise comparison of normalized array data from DUX4-fl vs GFP (left), and DUX4-s vs GFP (right). Blue dots: upregulated genes; red dots: downregulated genes; blue and red diagonal lines represent 2-fold change. Vertical and horizontal lines represent signal thresholds for calling genes present or absent. (B) Gene context of DUX4-fl binding sites (left) or MyoD binding sites (right) represented by peak density, adjusting for prevalence of gene context category in the genome. Promoter: +/−500 bp from the transcription start site (TSS); promoter.pro: +/−2 kb from the TSS; prime3: +/−500 nt from the end of the transcript; upstream: −2 kb to −10 kb upstream of the TSS; downstream: +2 kb to +10 kb from the end of the transcript; intergenic: >10 kb from any annotated gene. (C) Top: DUX4-fl motif Logo. The size of each nucleotide at a given position is proportional to the frequency of the nucleotide at that position, and the darkness of the line connecting two adjacent nucleotides represents corresponding dinucleotide frequency. Bottom: DUX4 binding motif matches MaLR repeat consensus sequence. We identified the best DUX4 binding sites (bracket) within the MaLR repeats annotated in the RepeatMasker track provided by the UCSC genome browser (hg18) and extended the motif in the flanking regions to reflect general MaLR repeat consensus.