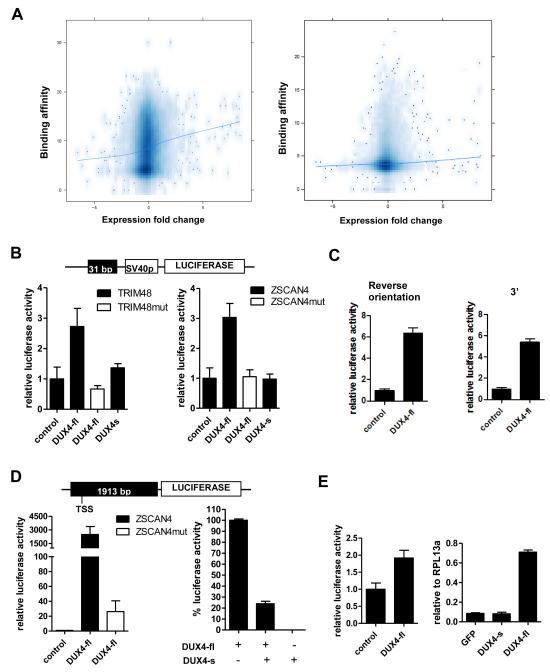
Figure 2. DUX4-fl activates transcription in vivo and DUX4-s can interfere with its activity.
(A) Comparison of DUX4 binding and regional gene transcription. The DUX4 peak height (Y-axis, square root transformation) of binding sites located within the CTCF-flanked domain of the TSS (left panel) or within +/− 2 KB of the TSS (right panel) plotted against the log-2 fold change of mRNA expression for the gene associated with the TSS (X-axis). The trend line shows a weak association between DUX4 binding and regional gene transcription. (B) Genomic fragments near theTRIM48 and ZSCAN4 genes containing DUX4 binding sites were cloned into pGL3-promoter reporter vector (schematic, top) and transfected into human rhabdomyoscaroma cell line RD. Cells were co-transfected with DUX4-fl or DUX4-s. pCS2-β galactosidase (beta-gal) was used to balance DNA amount in control condition. TRIM48mut and ZSCAN4mut, mutated binding sites. (C) DUX4-fl can act as an enhancer. Left: ZSCAN4 DUX4 binding site in reverse orientation relative to panel B upstream of the SV40 promoter. Right: ZSCAN4 DUX4 binding site in original orientation but moved 3′ downstream of the reporter gene. Luciferase activity set relative to control and error bars represent standard deviation of triplicates. (D) Left panel: Genomic fragment upstream of the ZSCAN4 translation start site containing four DUX4 binding sites was cloned into pGL3-basic luciferase vector (schematic, top). DUX4-fl highly activates the luciferase expression, whereas mutation of the binding sites (ZSCAN4mut) drastically reduces this induction. Right panel: Luciferase activity from DUX4-fl is set at 100% (pCS2-beta-gal is used to normalize the amount of plasmid). Co-transfection of equal amounts of DUX4-fl and DUX4-s diminishes luciferase activity. (E) Left: Genomic fragment from the LTR of THE1D MaLR element containing the DUX4 binding site were cloned into pGL3-promoter vector and tested for response to DUX4-fl as in (a). Right: Transcripts from endogenous retroelement MaLRs are upregulated by lentiviral transduction of DUX4-fl into primary human myoblasts. No upregulation is seen with lentiviral transduction of GFP or DUX4-s. Real-time RT-PCR quantitation is reported relative to internal standard RPL13a. All data represent mean +/− SD from at least triplicates.