Abstract
Injection of soluble cell signaling factors into degenerated intervertebral discs (IVDs) offers a minimally invasive treatment that could limit the processes of degeneration by stimulating native matrix repair. This study evaluated the regenerative capacity of degenerated nucleus pulposus (NP) cells obtained from patients undergoing anterior interbody fusions by measuring metabolic activity, DNA content, glycosaminoglycan (GAG) content, and cellular phenotype using qRT-PCR profiling with a custom array of 42 genes. NP cells were cultured in alginate for 7 days with 4 treatment groups: transforming growth factor beta 3 (TGFβ3) + dexamethasone (Dex), soluble factors released from notochordal cells (NCs) cultured in alginate (NCA), soluble factors released from NCs in their native tissue environment (NCT), and basal media. TGFβ3 + Dex stimulated degenerated human NP cells to proliferate and exhibit an anti-catabolic gene expression profile (with a decrease in ADAMTS5 and MMP1 compared to basal, and an increase in SOX9, decrease in ADAMTS5, MMP1, collagen I and collagen III compared to day 0), while NCA stimulated the greatest GAG per cell. We conclude that degenerated human NP cells exhibit regenerative potential, and that an optimal treatment will likely require treatments, such as TGFβ3 + Dex, which were able to increase cell metabolism and reduce catabolism, as well as treatments with factors found in NC conditioned medium, that were able to produce high amounts of GAG per cell. Additional studies to optimize NC culture conditions are required to determine if NC conditioned medium can be made with the capacity to enhance NP cell proliferation and metabolism.
Keywords: notochordal cells, TGFβ, intervertebral disc degeneration, cell therapy, nucleus pulposus
Degeneration of the intervertebral disc (IVD) is commonly a primary or secondary cause of low back pain.1 Current treatments, such as fusion and disc replacement, are aimed at relieving symptoms rather than restoring function. Injection of soluble cell signaling factors into degenerated IVDs is a promising minimally invasive treatment option with potential to slow or reverse the processes of disc degeneration by stimulating native matrix repair.2
The regenerative capacity of degenerated NP cells remains unclear as does the choice of biological stimulus. Degeneration of the nucleus pulposus (NP) is associated with substantial alterations in mechanical function associated with a decrease in aggrecan and collagen II production3 and an increase in degradation of aggrecan4 that limit the hydration capacity of the IVD’s extracellular matrix. Degeneration is also characterized by a loss of NP cells,5 an increase in cell death as well as cellular senescence,6 and an altered cellular phenotype; including a decrease in SOX9,7 and an increase in catabolic enzymes,8 and fibrotic collagens (collagen I and collagen III9). An effective treatment would therefore be defined as stimulating NP cells to increase glycosaminoglycan (GAG) production, cell number, and metabolic activity, as well as expression of genes characteristic of a non-degenerated NP phenotype (Table 1).
Table 1.
Criteria for Defining Proliferation, Metabolic Activity, Protein Production, and Phenotype for Non-Degenerated Human NP Cells
| Degenerative Changes in the NP | Goals for Effective Treatment as Measured in This Study |
|---|---|
| Loss of cells5 and increase in senescence6 | Increase in metabolic activity/DNA content |
| Cells with degenerative phenotype | Cells with nondegenerated phenotype |
| Decrease in collagen II and aggrecan production3 | Production of GAG, increase in ACAN and COL2A1 gene expression |
| Decrease in SOX97 | Increase in SOX9 |
| Increase in catabolic enzymes8 | Decrease in ADAMTS, MMPs |
| Increase in fibrotic collagens (collagen I and collagen III)9 | Decrease in fibrotic collagens (COL1A1 and COL3A1) |
While there are many candidate growth factors for injection into the IVD,2 this study focused on the hypothesis that repair will be achieved by recapitulating biological processes that occur during development. Notochordal cell (NC) conditioned media has been shown to increase proteoglycan production in bovine NP cells10,11 and to stimulate chondrocyte migration in a dose-dependent manner.12 This suggests that NCs, present throughout development, secrete soluble cell signaling factors that have potential therapeutic benefit. However, these studies used NCs in alginate to create conditioned media and it is also possible that NCs might release more or other soluble factors if they were cultured in native tissue. Therefore, conditioned media was created in two ways to compare the regenerative potential of NCs in alginate (NCA) with NCs in tissue (NCT).
Transforming growth factor beta 3 (TGFβ3) is another potential treatment, as its transcription is expressed during phases of morphogenesis in the IVD,13 suggesting it has an important role in development of the healthy IVD. TGFβ3 also regulated GAG and aggrecan synthesis in NP cells,14 stimulated human NP cells to create a proteoglycan and type II collagen-rich matrix,15 and in combination with dexamethasone (Dex); induced mesenchymal stem cells to adopt an IVD phenotype.16 Dex is also used in chondrogenic media and is an anti-inflammatory steroid shown to reduce the effects of IL-8 and TNFα on human NP cells.17 TGFβ3 + Dex, therefore, has potential to differentiate degenerated NP cells to a more highly anabolic non-degenerated NP phenotype.
The purpose of this study was to evaluate the regenerative potential of three treatments on degenerated human NP cells obtained from patients undergoing anterior interbody fusions. Characteristics of an effective treatment for disc degeneration involved the ability to increase NP cell metabolic activity, proliferation, GAG production, and stimulate a non-degenerated NP phenotype as measured by gene expression profiling. It was hypothesized that degenerated NP cells would have regenerative potential, and that media from NC conditioned media would offer promise to enhance GAG production and be an effective treatment. Should NC conditioned media be a useful stimulant for NP cells and eventually IVDs in vivo, then isolation of the important cytoactive factors found in NC conditioned media could be identified and synthesized for therapeutic injection or other intervention.
METHODS
Generation of NC Conditioned Medium (NCA and NCT)
Porcine spines (2- to 8-month-old within 24 h of death, Animal facility Research 87 Inc, Boylston, MA) were aseptically dissected to remove NC-rich NP tissue. The NP tissue was either soaked directly in conditioning media (CM; low glucose DMEM, 1% penicillin/streptomycin, 0.05% Fungizone, and 2% 5 M NaCl/0.4 M KCl) 3 IVDs per 30 mls media to create NCT, or digested (0.2% protease for 1 h, 0.025% collagenase for 18 h, non-enzymatic cell dissociation solution for 2 h, all from Sigma–Aldrich, St. Louis, MO) to release the cells.18 The cells were then counted on a hemocytometer (average ratio of NC:NP cells was 88%:12%) and re-suspended in alginate beads at a density of 2 × 106 cells/ml of alginate in CM to create NCA. This density was chosen to ensure a similar density of cells per ml of CM as NCT which had ~2 million NC/NP cells per disc explant (20,000 cells/ml of CM). At 2 × 106 cells/ml of alginate there are ~ 40,000 cells/single bead and 10 beads in a well with 2 ml CM (20,000 cells/ml of CM). Both NCT and NCA were incubated for 4 days in hypoxia (5% O2, 5% CO2, 37°C)18 and filtered (3K Amicon Ultra-15, Millipore, Billerica, MA). The retained filtrate was re-suspended in basal (CM with 1% insulin–transferrin–selenium; Sigma–Aldrich) to create the final NCT and NCA media.
Isolation and Culture of Human NP Cells
Human NP tissue was obtained from patients undergoing anterior interbody fusions for low back pain secondary to degenerative disc disease with institutional review board approval and graded by the surgeon as either Grade IV (n = 2, female age 51 and male age 58) or Grade V (n = 1, female age 47) on the Thompson scale. The cells were released from the NP tissue by sequential enzymatic digestion (0.2% protease, 1 h; 0.2% collagenase, 4 h, both from Sigma–Aldrich). Isolated cells were filtered through a 70-µm mesh (Fisher, Pittsburgh, PA) and washed twice with PBS. They were expanded in culture in high glucose DMEM containing 10% fetal bovine serum, 1% pen/strep, 0.05% fungi-zone, and 50 µg/ml ascorbate. After expansion (passage II), human NP cells were counted with a hemocytometer and were either used directly for gene expression analysis (day 0, normalization for the ΔΔCt method) or rinsed in 0.15 M NaCl and encapsulated in low viscosity alginate (Sigma–Aldrich) at a density of 2 × 106 cells/ml of 1.2% alginate. For each group alginate beads without cells were also created. Alginate constructs were cultured in 12-well plates (10 beads/well) for 7 days in hypoxia, with two media changes. Media conditions included basal (CM + 1% insulin–transferrin–selenium; Sigma–Aldrich), TGFβ3 + Dex (basal media with 10 ng/ml TGFβ319 and 10% Dex16), NCA or NCT. Analysis of metabolic activity, DNA content, GAG content, and gene expression were performed post-culture.
Dependent Variables
The MTT ((4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was used to determine metabolic activity.20 Human NP cells in alginate constructs were incubated in a 2 mg/ml solution of MTT21 in DMEM for 4 h. The alginate was dissolved (55 mM sodium citrate, 30 mM EDTA, 0.15 M NaCl) for 10 min. The cells’ suspension was then centrifuged, supernatant removed, cell pellet lysed with 200 µl of DMSO, and read (570 nm).
To determine DNA content and GAG, the alginate constructs were dissolved, centrifuged, and lysed with 200 µl RNeasy Lysis buffer (Qiagen, Valencia, CA). DNA content (cell number) of samples was analyzed using the Picogreen dsDNA quantification kit (Invitrogen) from the lysate (10 µl, in duplicate) according to the manufacturer’s instructions. GAG was measured in the construct (lysate), media, and in the alginate supernatant using the dimethyl methylene blue (DMMB) assay.22 The total GAG includes the total amount in the construct, media, and supernatant, and this quantity was adjusted by subtracting from the experimental values the amount of GAG detected in cell-free constructs (i.e., cell-free alginate constructs in identical media conditions for identical experimental durations) to account for any GAG that may be present in the conditioned media or the effect of charged alginate residue on the DMMB assay. We interpret this as GAG produced, and further note that GAG in media and supernatant (<0.7 and <0.02 µg/ml, respectively) were small or negligible when compared to the amount measured in the construct.
To define a non-degenerated NP gene expression profile a custom PCR array with 42 genes per sample was used to evaluate a combination of anabolic, catabolic, anti-catabolic, pro-inflammatory cytokines, and various growth factor genes that are known to change with degeneration. Gene expression was assessed by dissolving the alginate to release the cells. The cells were washed twice with the dissolving buffer, lysed with 300 µl RNeasy Lysis buffer (Qiagen), and stored at −80°C (1 µl RNAse inhibitor). RNA was extracted, cDNA synthesized, and custom RT2 Profiler™ SYBR green PCR arrays (CAPH-0817A; SABiosciences, Frederick, MD) were run (Supplement 1 and Ref. 23). Relative gene expression was calculated with the comparative Ct method normalized to the gene expression of cells before culture and 3 housekeeping genes (18SrRNA, GAPDH, and ACTB). For normalization purposes, undetermined values for day 0 were given an arbitrary value of 40 (genes that were not expressed in some of the patients at day 0: COL2A1, TIMP3, and TNFα).
Statistics
All data were assessed for normality using the Ryan–Joiner test. A one-way ANOVA was used to assess differences between groups for MTT, Picogreen, DMMB, and qRT-PCR, with a Fisher’s PLSD test to evaluate which groups were significantly different. One sample t-tests were done on ΔΔCt values to assess differences from day 0 (ΔΔCt = 0). All statistics were performed with Minitab software with significance defined as p < 0.05.
RESULTS
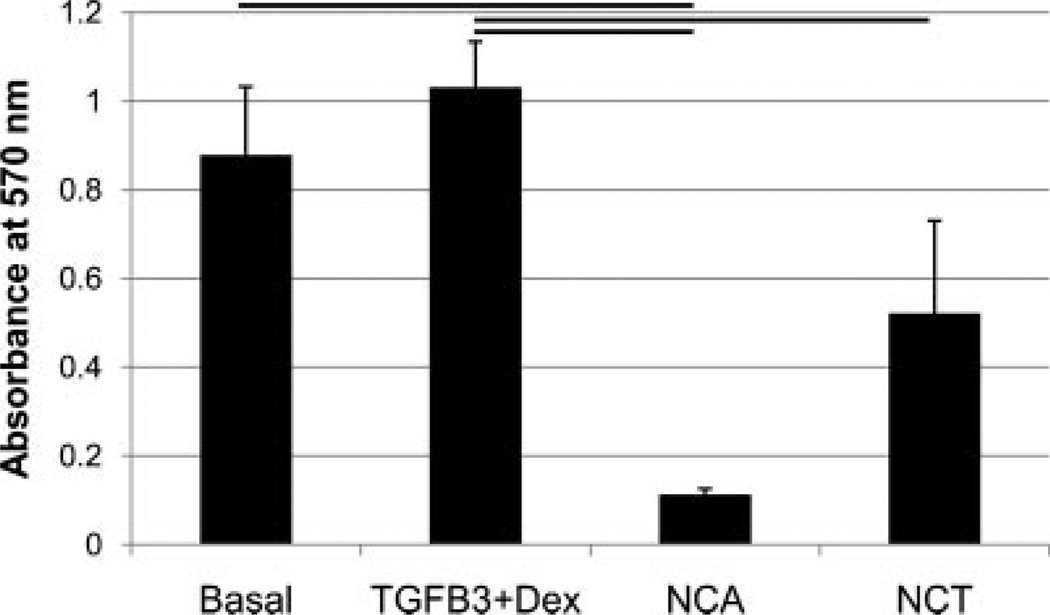
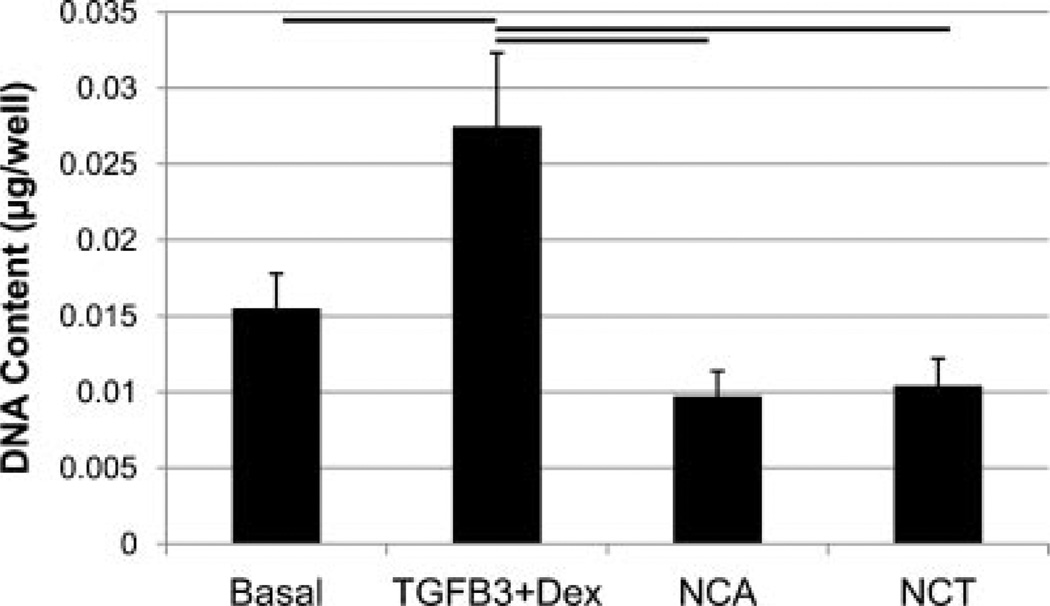
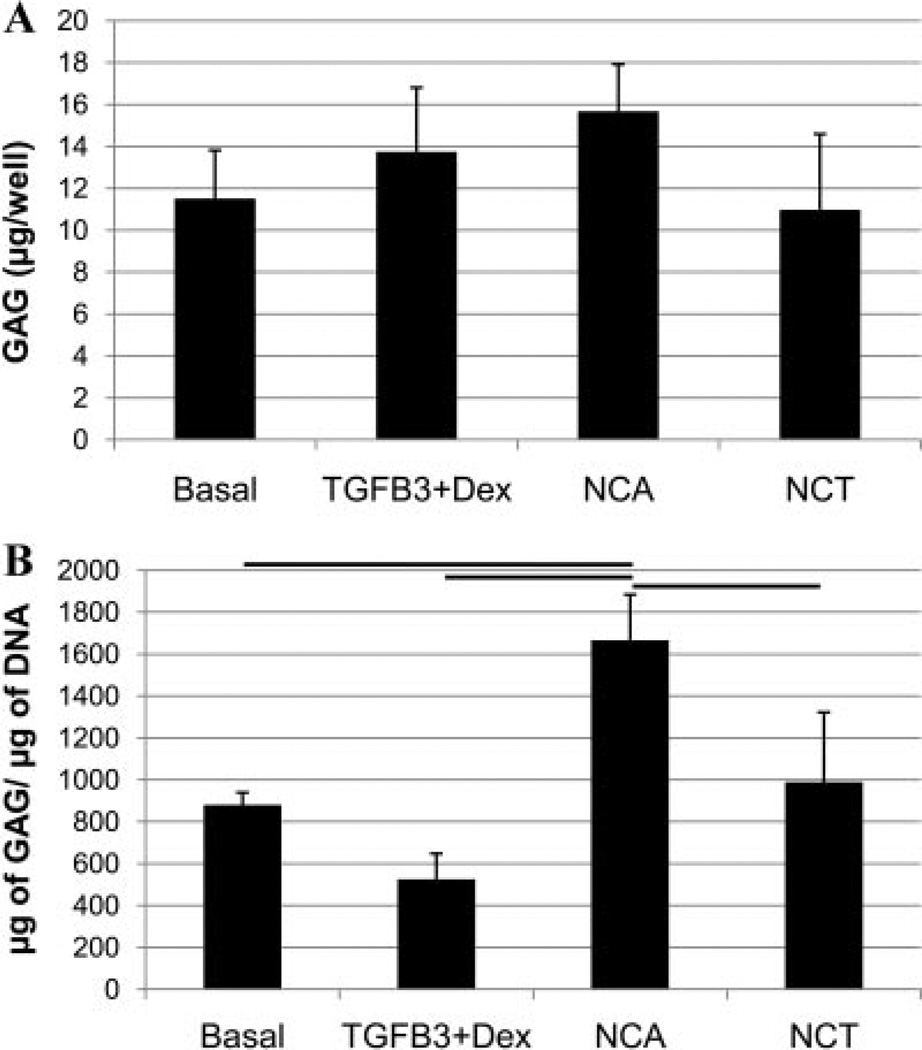
The MTT assay indicated that human NP cells cultured in TGFβ3 + Dex media demonstrated a similar metabolic activity as cells cultured in the basal control group (Fig. 1), however, cells cultured in NCA and NCT media had 90% and 50% less MTT absorbance than TGFβ3 + Dex, respectively (p < 0.05). There was significantly more DNA in wells treated with TGFβ3 + Dex, compared to NCA, NCT, and the basal group (Fig. 2). There were no significant differences in the amount of total GAG per well between groups (Fig. 3A); however, there was significantly more GAG per cell for the NCA group compared to the other groups (Fig. 3B).
Figure 1.
The MTT assay was used to determine the cell’s metabolic activity. Human NP cells cultured in TGFβ3 + Dex media demonstrated a similar metabolic activity as cells cultured in the basal control group, with cells cultured in NCA and NCT media 90% and 50% less than TGFβ3 + Dex, respectively. Lines indicated significant difference of p < 0.05, error bars represent SEM.
Figure 2.
DNA content (or cell number) of samples was analyzed using the Picogreen dsDNA quantification kit. Bar indicates significance of p < 0.05 and error bars are SEMs. There was significantly more DNA with media containing TGFβ3 + Dex, compared to NCA and NCT, consistent with MTT results.
Figure 3.
DMMB analysis was used to assess the amount of GAG in the alginate constructs. Overall there were no significant changes between groups per well (A); however, there was significantly more GAG per cell (B) with the NCA treatment compared to the other groups. Lines indicate significance of p < 0.05, and error bars are SEMs.
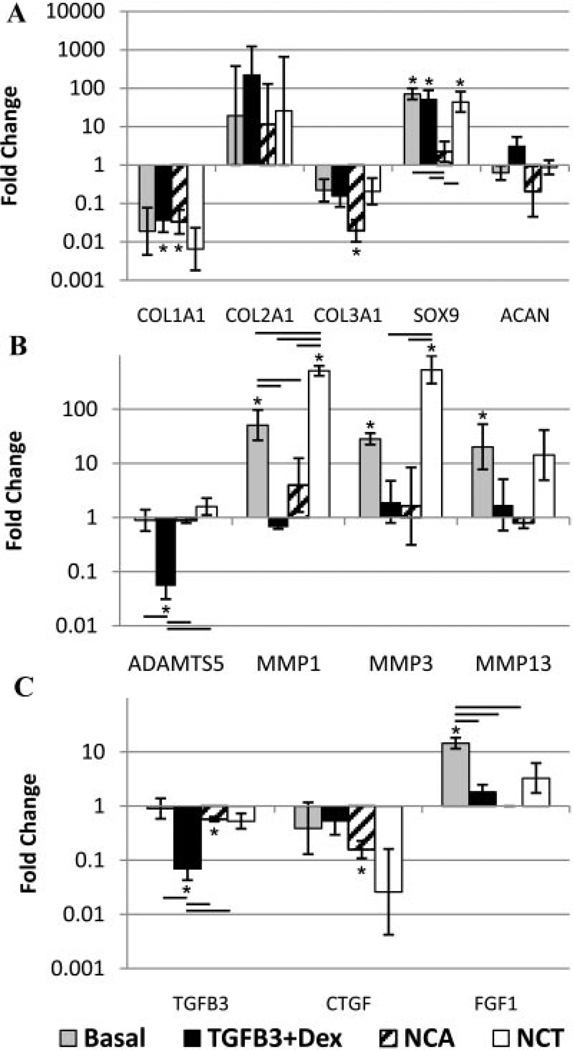
Gene expression (Fig. 4) showed a general downregulation of fibrotic genes, collagen I, and collagen III, and a general upregulation of collagen II for all treatment groups compared to day 0 (with no differences from the basal control). The TGFβ3 + Dex group had an increasing trend of aggrecan, as well as a significant increase in SOX9 from day 0. An increase in SOX9 was also observed for cells cultured in basal and NCT media with the only significant change from basal being a downregulation in the NCA group. All of the groups showed no significant changes in collagen X expression (Supplement 1 for complete gene expression data). Catabolic gene expression was significantly upregulated for MMP1 (from day 0 and basal) and MMP3 (from day 0) for the NCT group, while ADAMTS5 and MMP1 were significantly downregulated for cells cultured in TGFβ3 + Dex (from day 0 and basal). There was a significant decrease in CTGF for the NCA group from day 0 that was not significantly different from basal, and TGFβ3 for the TGFβ3 + Dex group (significantly down from day 0 and compared to the basal control).
Figure 4.
qRT-PCR results for human degenerated NP cells. Fold change in mRNA levels were calculated with the ΔΔCt method relative to three housekeeping genes and human degenerated NP cells before culture from the same patient. *Significantly different from day 0 (p < 0.05), line indicates differences between groups (p < 0.05), and error bars are SEMs. (A) Anabolic gene expression: a general downregulation of collagen I and collagen III for all treatment groups. Basal, TGFβ3 + Dex, and NCT significantly increased Sox 9. (B) Catabolic gene expression: TGFβ3 + Dex treatment significantly downregulated ADAMTS5 and MMP1, while NCT significantly upregulated MMP1 and MMP3. Basal significantly upregulated MMP1, MMP3, and MMP13, with no changes in MMPs for NCA. (C) Growth factor gene expression: TGFβ3 + Dex resulted in a significant decrease in TGβ3, while NCA caused a significant decrease in CTGF. FGF1 was significantly upregulated in the basal group.
DISCUSSION
This screening study evaluated the effectiveness of three treatments on degenerated human NP cells isolated from IVD tissue from patients undergoing anterior interbody fusions to evaluate their potential to regenerate or slow IVD degeneration. Treatment effectiveness was defined by its ability to increase metabolic activity of the cells (MTT), cell numbers (picogreen), and GAG content (DMMB), and to promote a non-degenerated NP phenotype based on gene expression profiling. In support of the hypothesis, degenerated NP cells were responsive to soluble factor treatments demonstrating potential for therapeutic interventions, and the NCA group promoted the greatest GAG per cell. However, TGFβ3 + Dex showed regenerative potential as well by stimulating degenerated human NP cells to proliferate and exhibit a gene expression profile more characteristic of an anti-catabolic NP phenotype (decrease in ADAMTS5 and MMP1 from basal and a decrease in ADAMTS5, MMP1, collagen I, and collagen III with an increase in SOX9 compared to day 0).
TGFβ3 + Dex significantly enhanced metabolic activity (MTT assay) and cell proliferation (DNA content) within the alginate constructs compared to NCA and NCT, countering the decrease in cell number and increase in cellular senescence that occurs with degeneration. 5,6 TGFβ3 + Dex also induced an anti-catabolic NP phenotype at the gene level. There was a significant downregulation of ADAMTS5 and MMP1 compared to both day 0 and basal conditions which are responsible for degrading matrix proteins in the degenerated IVD.24
The SOX9 transcription factor which directly regulates the collagen II gene,25 was upregulated in the basal, TGFβ3 + Dex, and NCT group (with the only significant difference from basal being a downregulation in the NCA group). The collagen II gene displayed an upward trend for all groups with no difference between groups, although this was not significant. It is expected that expression may become significantly increased after longer time points as the SOX9 gene gets transcribed to protein. In this study, there was also a general downregulation of collagen I and collagen III gene expression with all treatments. As these changes occurred in all of the groups, including the basal control, it is clear that the transition to 3D alginate culture conditions alone can alter gene expression.
Conditioning of degenerated NP cells with NCA resulted in a similar amount of GAG per well despite having fewer number of cells than the TGFβ3 + Dex group. This translated to significantly more GAG per cell than the other treatment groups, supporting studies with bovine cells cultured in NC conditioned media.10,11 Although NCA stimulated maximal GAG per cell, both NC conditioned groups (NCT and NCA) had a fewer number of cells (Picogreen) and a significant decrease in the metabolic activity (MTT assay) of the cells compared to TGFβ3 + Dex. This is in contrast to the prior findings with bovine cells that indicated a fourfold increase in cell proliferation with NC conditioned media.11 In addition NCT resulted in a catabolic shift in gene expression which is more consistent with degeneration where there are decreased numbers of cells and an increase in catabolism.5,24 The observed effects with NC conditioning suggests that either the secreted soluble factors require an optimal dose that was not achieved, or that NC conditioned media is cytotoxic and may not secrete the factors required to stimulate human degenerated NP cells to proliferate and express genes more consistent with a non-degenerated phenotype. Additional studies to optimize NC culture conditions are required to determine if NC conditioned medium can be made with the capacity to enhance NP cell proliferation and metabolism.
Evidence of a possible autocrine (negative) feedback response was observed for growth factor and matricellular gene expression in all of the groups. Cells cultured in TGFβ3 + Dex downregulated TGFβ3, while both NC groups downregulated CTGF (significant for NCA) from day 0, a matricellular protein produced by NCs.26 While the differences in CTGF expression from the other groups are only trends, the results suggest that additional investigations of matricellular proteins may be important in addition to growth factors studies. For example, CTGF which binds to aggrecan and is known to modulate extracellular matrix production,27 and could be a contributor to maintenance of similar levels of GAG per well for NC groups compared to TGFβ3 + Dex, despite the decreased number of cells.
There are many candidate growth factors for injection into the IVD.2 A number of studies have examined their effect on animal models, and while there are many advantages to these experiments, they may not translate to human degenerated cells. Few studies have used human cells obtained from surgery to examine the effects of growth factors,15,28–31 despite the clinical relevance. While some of these growth factors have stimulated human cells to increase GAG and collagen II synthesis,28,30 they were associated with an increase in type I collagen, indicating a more fibrotic phenotype that was not demonstrated in this study compared to day 0 or from basal for any of the groups.
Some limitations in this study are noteworthy. A healthy NP has a ratio of 27:1 proteoglycan to collagen content32; therefore collagen I and collagen II ELISA were performed on the media to determine a ratio. Since on average only ~14 µg of GAG were present in each well following 7 days of culture, it is not surprising that the collagen content was below sensitivity of the ELISA (0.05 µg) so that longer culture durations are required to obtain detectable measurements of collagen. The use of three human subjects limits the statistical power of this study so that it is difficult to make conclusions when significant differences were not found. However, the three patients showed similar trends, displayed significant differences between groups, and exhibited conclusive findings so that this sample size of three patients was sufficient to achieve the goals of this study. The basal group was not used to determine baseline mRNA levels since it does not represent a realistic baseline treatment option, altered mRNA expression compared to day 0 levels with increased gene expression of MMP1, MMP3, MMP13, TIMP3, FGF1, and SOX9), and because the increase in FGF1 suggests some differentiation. Consequently, qRT-PCR levels of the day 0 (i.e., the beginning of alginate culture) group were used as a baseline to determine how treatments affected the gene expression relative to the degenerated state. When discussing GAG in the constructs, MTT, and Picogreen the relevant comparisons were still between groups. Human NCs disappear within the first decade of life,33 making a human source of NC cells very limited. Therefore, Porcine NCs were chosen based on their availability in a large animal IVD. Degenerated human NP cells are the most clinically relevant and were used in this study. Consequently, a single species study was not feasible, and we investigated this xenogenic model due to its greater clinical relevance. Previous studies have used xenogenic models to examine the effect of canine NC conditioned media on bovine NP cells,10,11,26 as well as porcine NC conditioned media with human MSCs,23,34 making this model a reasonable choice.
In conclusion, TGFβ3 + Dex showed the greatest regenerative potential of the treatments tested to stimulate degenerated NP cells to proliferate and express a gene expression profile more characteristic of an anti-catabolic phenotype; while NCA showed the greatest potential to increase GAG synthesis per cell. The high GAG content may be at least partly associated with the presence of CTGF or other matricellular proteins found in NC conditioned media. However, the NC conditioned media lacked anti-catabolic or proliferative capacity which could be due to un-optimized NC culture conditions. It is likely that an optimal treatment will combine mitogenic/anti-catabolic agents such as TGFβ3 + Dex (to increase cell metabolism and promote an anti-catabolic phenotype) with cyto-active constituents of NC conditioned media (to enhance GAG production per cell). While these results are promising and provide a rapid screening of three different treatments associated with recapitulation of development, a longer study is necessary to determine if these results are sustained, and if in vitro simulations of human degenerated NP cells will translate to a clinically significant treatment in vivo with symptom modifying and structurally modifying effects. Targeting certain degrees of disc degeneration as well as endplate structure, viability, and metabolic activity of the cells are likely to be crucial steps.35 Grades IV and V human NP cells used in this study are often considered a more advanced degeneration level than is optimal for regenerative therapies, and future work should include determining if grade of degeneration is an important factor in responsiveness of NP cells to biological therapies. It also has to be determined whether there are sufficient cells in the degenerated NP to make injection of therapeutics an effective treatment option and whether the poor nutrient supply can withstand an increase in metabolic activity of the cells.36 We conclude that injection of soluble factors found in NC conditioned media and TGFβ3 + Dex may offer promise to stimulate degenerated NP cells and that measurements of cell proliferation, metabolic activity, proteins produced, and phenotypic profiling are important evaluations of therapeutic potential.
Supplementary Material
ACKNOWLEDGMENTS
This work supported by grants from the NIH (R21AR056037) and by the AO Research Fund (project F-09-10I) of the AO Foundation. We gratefully acknowledge technical assistance of Tim Hunter and Mary Lou Shane in the Vermont Cancer Center DNA Analysis Facility and helpful discussions with Dr. David Brigstock regarding CTGF.
Footnotes
Additional supporting information may be found in the online version of this article.
REFERENCES
- 1.Cheung KM, Karppinen J, Chan D, et al. Prevalence and pattern of lumbar magnetic resonance imaging changes in a population study of one thousand forty-three individuals. Spine (PhilaPa 1976) 2009;34:934–940. doi: 10.1097/BRS.0b013e3181a01b3f. [DOI] [PubMed] [Google Scholar]
- 2.Masuda K, An HS. Prevention of disc degeneration with growth factors. Eur Spine J. 2006;15 Suppl 3:S422–S432. doi: 10.1007/s00586-006-0149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniou J, Steffen T, Nelson F, et al. The human lumbar intervertebral disc—evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J Clin Investig. 1996;98:996–1003. doi: 10.1172/JCI118884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verzijl N, DeGroot J, Bank RA, et al. Age-related accumulation of the advanced glycation endproduct pentosidine in human articular cartilage aggrecan: the use of pentosidine levels as a quantitative measure of protein turnover. Matrix Biol. 2001;20:409–417. doi: 10.1016/s0945-053x(01)00158-5. [DOI] [PubMed] [Google Scholar]
- 5.Gruber HE, Hanley EN., Jr Analysis of aging and degeneration of the human intervertebral disc. Comparison of surgical specimens with normal controls. Spine (PhilaPa 1976) 1998;23:751–757. doi: 10.1097/00007632-199804010-00001. [DOI] [PubMed] [Google Scholar]
- 6.Le Maitre CL, Freemont AJ, Hoyland JA. Accelerated cellular senescence in degenerate intervertebral discs: a possible role in the pathogenesis of intervertebral disc degeneration. Arthritis Res Ther. 2007;9:R45. doi: 10.1186/ar2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gruber HE, Norton HJ, Ingram JA, et al. The SOX9 transcription factor in the human disc: decreased immunolocalization with age and disc degeneration. Spine (PhilaPa 1976) 2005;30:625–630. doi: 10.1097/01.brs.0000155420.01444.c6. [DOI] [PubMed] [Google Scholar]
- 8.Le Maitre CL, Freemont AJ, Hoyland JA. Localization of degradative enzymes and their inhibitors in the degenerate human intervertebral disc. J Pathol. 2004;204:47–54. doi: 10.1002/path.1608. [DOI] [PubMed] [Google Scholar]
- 9.Boos N, Nerlich AG, Wiest I, et al. Immunolocalization of type X collagen in human lumbar intervertebral discs during ageing and degeneration. Histochem Cell Biol. 1997;108:471–480. doi: 10.1007/s004180050187. [DOI] [PubMed] [Google Scholar]
- 10.Aguiar DJ, Johnson SL, Oegema TR. Notochordal cells interact with nucleus pulposus cells: regulation of proteoglycan synthesis. Exp Cell Res. 1999;246:129–137. doi: 10.1006/excr.1998.4287. [DOI] [PubMed] [Google Scholar]
- 11.Erwin WM, Inman RD. Notochord cells regulate intervertebral disc chondrocyte proteoglycan production and cell proliferation. Spine (PhilaPa 1976) 2006;31:1094–1099. doi: 10.1097/01.brs.0000216593.97157.dd. [DOI] [PubMed] [Google Scholar]
- 12.Kim KW, Ha KY, Lee JS, et al. Notochordal cells stimulate migration of cartilage end plate chondrocytes of the intervertebral disc in in vitro cell migration assays. Spine J. 2009;9:323–329. doi: 10.1016/j.spinee.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 13.Pelton RW, Dickinson ME, Moses HL, et al. In situ hybridization analysis of TGF beta 3 RNA expression during mouse development: comparative studies with TGF beta 1 and beta 2. Development. 1990;110:609–620. doi: 10.1242/dev.110.2.609. [DOI] [PubMed] [Google Scholar]
- 14.Hiyama A, Gogate SS, Gajghate S, et al. BMP-2 and TGF-beta stimulate expression of beta 1,3-glucuronosyl transferase 1 (GlcAT-1) in nucleus pulposus cells through AP1, TonEBP, and Sp1: role of MAPKs. J Bone Miner Res. 2010;25:1179–1190. doi: 10.1359/jbmr.091202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haberstroh K, Enz A, Zenclussen ML, et al. Human intervertebral disc-derived cells are recruited by human serum and form nucleus pulposus-like tissue upon stimulation with TGF-beta3 or hyaluronan in vitro. Tissue Cell. 2009;41:414–420. doi: 10.1016/j.tice.2009.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Steck E, Bertram H, Abel R, et al. Induction of intervertebral disc-like cells from adult mesenchymal stem cells. Stem Cells. 2005;23:403–411. doi: 10.1634/stemcells.2004-0107. [DOI] [PubMed] [Google Scholar]
- 17.Jee BK, Surendran S, Park KM, et al. Role of tumor necrosis factor-alpha, interleukin-8, and dexamethasone in the focal adhesion kinase expression by human nucleus pulposus cells. Spine (PhilaPa 1976) 2007;32:30–35. doi: 10.1097/01.brs.0000250997.24617.a4. [DOI] [PubMed] [Google Scholar]
- 18.Guehring T, Wilde G, Sumner M, et al. Notochordal intervertebral disc cells: sensitivity to nutrient deprivation. Arthritis Rheum. 2009;60:1026–1034. doi: 10.1002/art.24407. [DOI] [PubMed] [Google Scholar]
- 19.Risbud MV, Di Martino A, Guttapalli A, et al. Toward an optimum system for intervertebral disc organ culture: TGF-beta 3 enhances nucleus pulposus and anulus fibrosus survival and function through modulation of TGF-beta-R expression and ERK signaling. Spine (PhilaPa 1976) 2006;31:884–890. doi: 10.1097/01.brs.0000209335.57767.b5. [DOI] [PubMed] [Google Scholar]
- 20.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 21.Chen J, Yan W, Setton LA. Static compression induces zonal-specific changes in gene expression for extracellular matrix and cytoskeletal proteins in intervertebral disc cells in vitro. Matrix Biol. 2004;22:573–583. doi: 10.1016/j.matbio.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 22.Farndale RW, Buttle DJ, Barrett AJ. Improved quantitation and discrimination of sulphated glycosaminoglycans by use of dimethylmethylene blue. Biochim Biophys Acta. 1986;883:173–177. doi: 10.1016/0304-4165(86)90306-5. [DOI] [PubMed] [Google Scholar]
- 23.Purmessur D, Schek RM, Abbott RD, et al. Notochordal conditioned media from tissue increases proteoglycan accumulation and promotes a healthy nucleus pulposus phenotype in human mesenchymal stem cells. Arthritis Res Ther. 2011;13:R81. doi: 10.1186/ar3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le Maitre CL, Pockert A, Buttle DJ, et al. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35 doi: 10.1042/BST0350652. 652-L 655. [DOI] [PubMed] [Google Scholar]
- 25.Bell DM, Leung KK, Wheatley SC, et al. SOX9 directly regulates the type-II collagen gene. Nat Genet. 1997;16:174–178. doi: 10.1038/ng0697-174. [DOI] [PubMed] [Google Scholar]
- 26.Erwin WM, Ashman K, O’Donnel P, et al. Nucleus pulposus notochord cells secrete connective tissue growth factor and up-regulate proteoglycan expression by intervertebral disc chondrocytes. Arthritis Rheum. 2006;54:3859–3867. doi: 10.1002/art.22258. [DOI] [PubMed] [Google Scholar]
- 27.Aoyama E, Hattori T, Hoshijima M, et al. N-terminal domains of CCN family 2/connective tissue growth factor bind to aggrecan. Biochem J. 2009;420:413–420. doi: 10.1042/BJ20081991. [DOI] [PubMed] [Google Scholar]
- 28.Yang SH, Lin CC, Hu MH, et al. Influence of age-related degeneration on regenerative potential of human nucleus pulposus cells. J Orthop Res. 2010;28:379–383. doi: 10.1002/jor.20988. [DOI] [PubMed] [Google Scholar]
- 29.Le Maitre CL, Freemont AJ, Hoyland JA. Expression of cartilage-derived morphogenetic protein in human intervertebral discs and its effect on matrix synthesis in degenerate human nucleus pulposus cells. Arthritis Res Ther. 2009;11:R137. doi: 10.1186/ar2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DJ, Moon SH, Kim H, et al. Bone morphogenetic protein-2 facilitates expression of chondrogenic, not osteogenic, phenotype of human intervertebral disc cells. Spine (PhilaPa 1976) 2003;28:2679–2684. doi: 10.1097/01.BRS.0000101445.46487.16. [DOI] [PubMed] [Google Scholar]
- 31.Kim H, Lee JU, Moon SH, et al. Zonal responsiveness of the human intervertebral disc to bone morphogenetic protein-2. Spine (PhilaPa 1976) 2009;34:1834–1838. doi: 10.1097/BRS.0b013e3181ae18ba. [DOI] [PubMed] [Google Scholar]
- 32.Mwale F, Roughley P, Antoniou J. Distinction between the extracellular matrix of the nucleus pulposus and hyaline cartilage: a requisite for tissue engineering of intervertebral disc. Eur Cell Mater. 2004;8:58–63. doi: 10.22203/ecm.v008a06. discussion 63–64. [DOI] [PubMed] [Google Scholar]
- 33.Phelip X. Why the back of the child? Eur Spine J. 1999;8:426–428. doi: 10.1007/s005860050200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Korecki CL, Taboas JM, Tuan RS, et al. Notochordal cell conditioned medium stimulates mesenchymal stem cell differentiation toward a young nucleus pulposus phenotype. Stem Cell Res Ther. 2010;1:18. doi: 10.1186/scrt18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An HS, Thonar EJ, Masuda K. Biological repair of intervertebral disc. Spine (PhilaPa 1976) 2003;28:S86–S92. doi: 10.1097/01.BRS.0000076904.99434.40. [DOI] [PubMed] [Google Scholar]
- 36.Urban JP, Smith S, Fairbank JC. Nutrition of the intervertebral disc. Spine (PhilaPa 1976) 2004;29:2700–2709. doi: 10.1097/01.brs.0000146499.97948.52. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.