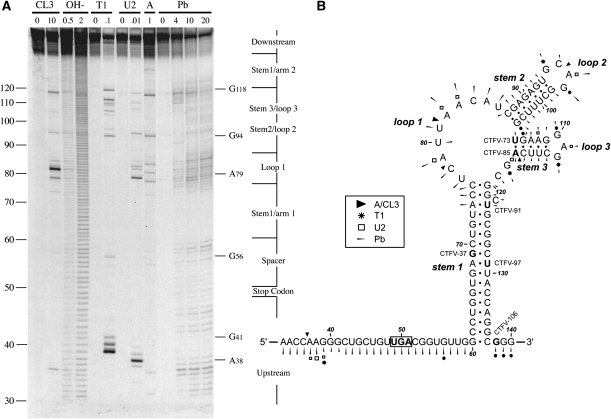
FIGURE 2.
Structure probing of the CTFV readthrough signal. (A) RNA derived by transcription of pCTFV-121-T3/BamHI with T3 RNA polymerase was 5′ end-labeled with [γ-33P]ATP and subjected to limited RNase or chemical cleavage using structure-specific probes. Sites of cleavage were identified by comparison with a ladder of bands created by limited alkaline hydrolysis of the RNA (OH-; RNA heated to 100°C for 0.5 or 2 min) and the position of known RNase U2 and T1 cuts, determined empirically. Products were analyzed on a 10% acrylamide/7M urea gel containing formamide. Enzymatic structure probing was with RNases CL3, T1, U2, and A. Uniquely cleaved nucleotides were identified by their absence in untreated control lanes (0). The number of units of enzyme added to each reaction is indicated. Chemical structure probing was with lead acetate (Pb; mM concentration in reaction). (B) The sequence of the probed CTFV RNA and the inferred secondary structure. The sensitivity of bases in the CTFV readthrough region to the various probes is shown for an mfold prediction. The first base of the transcript is numbered 1. The reactivies of the T1 (asterisk), U2 (open square), A and CL3 (black triangle) probes are marked. Lead cleavages are indicated by thin arrows. The size of the symbols is approximately proportional to the intensity of cleavage at that site. Also indicated is the location of the 3′ edge of the truncated versions of CTFV-307 (labeled as in Fig. 1), with the last viral base in each truncation emboldened.