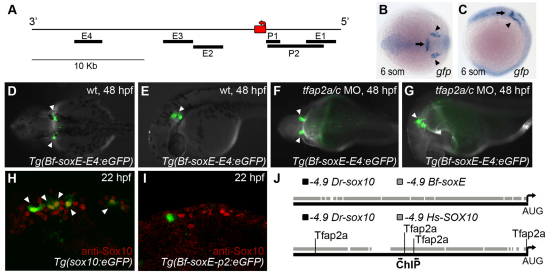
Fig. 6.
Amphioxus soxE elements fail to drive neural crest specific expression in zebrafish embryos. (A) Schematic of amphioxus soxE locus showing location of DNA elements tested in zebrafish reporter analysis. Red box, coding region of soxE; red arrow, initial AUG. Genomic coordinates are shown in supplementary material Table S2. (B,C) Anterior dorsal (B) and lateral (C) views of a Tg(Bf-soxE-E4:eGFP) zebrafish embryo fixed at 12 hpf and processed for gfp expression. gfp expression in midbrain-hindbrain (arrows) and otic placodes (arrowheads) is apparent. (D-G) Dorsal (D,F) and lateral (E,G) views of live, 48 hpf Tg(Bf-soxE-E4:eGFP) embryos. Arrowheads in D and E indicate GFP expression in otic vesicle (observed in five of 20 injected embryos). (F,G) A Tg(Bf-soxE-E4:eGFP) embryo injected with MOs targeting tfap2a and tfap2c, revealing eGFP expression in otic vesicle (arrowheads) despite an absence of melanophores (otic vesicle expression seen in three of 14 similarly injected transgenic embryos). (H,I) Lateral views of 22 hpf fixed embryos injected with (H) sox10:egfp plasmid (Wada et al., 2005), showing many GFP-positive, anti-Sox10-positive NC cells (arrowheads) in the dorsum (18/22 injected embryos showed dorsal GFP-positive cells), or (I) an analogous plasmid harboring 4.9 kb upstream of amphioxus soxE (Bf-soxE-p2:eGFP), with very few such cells (4/22 embryos showed few dorsal GFP-positive cells all of which were negative for anti-Sox10 immunoreactivity). (J) Modified output of ConSite software showing conservation of sequence and Tfap2 transcription factor binding sites in 4.9 kb of genomic DNA upstream of the start of the initiator AUG in human SOX10 (–4.9 Hs-SOX10) and zebrafish sox10 (–4.9 Dr-sox10). Arrows, region tested in ChIP analysis. An identical comparison of amphioxus soxE (–4.9 Bf-soxE) and zebrafish sox10 using identical parameters fails to detect conserved Tfap2 binding sites. Embryos shown with anterior to the left, unless otherwise indicated.