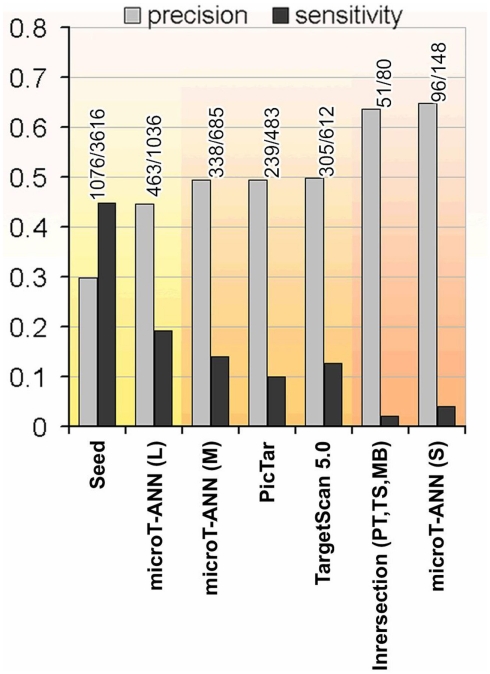
Figure 3.
The precision and sensitivity of representative target prediction methods tested on the proteomics data. The tested programs are: Pictar, TargetScan 5.0, and DIANA-microT-ANN using the suggested three different score cutoffs denoted as strict (S), medium (M), and loose (L). Additionally the performance of a seed occurrence measure, detecting genes containing at least one occurrence of the miRNA seed is shown as a very sensitive prediction criterion, and the commonly used intersection of the predicted targets of Pictar, TargetScan 5.0, and miRBase as a very strict and precise measure. Three groups are highlighted indicating the methods with high sensitivity, medium, and high precision (from left to right).