Figure 4.
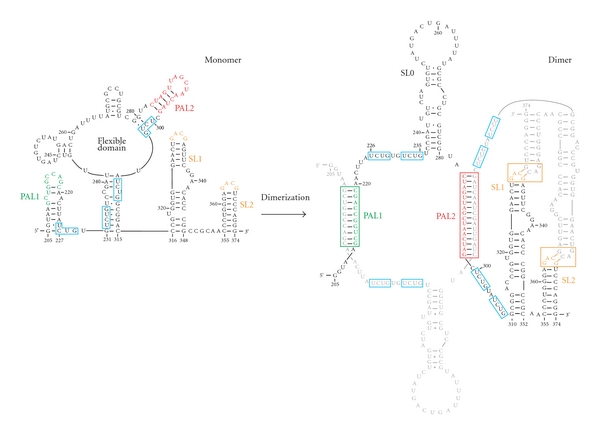
The Moloney MLV dimerization/packaging signal. The figure shows the secondary structure of the 170-base “minimal dimerization active sequence” (nt 205–374) [19] in both monomeric and dimeric forms. Two palindromic sequences, “PAL1” (green) and “PAL2” (red), are contained within stem loops in the monomer but open out and pair intermolecularly in the dimer. The two monomers are also connected in the dimer by base pairing between the “CG” moieties in the “GACG” loops of a pair of stem loops (“SL1” and “SL2”, orange). The RNA also contains two motifs with the sequence UCUG-UPu-UCUG (blue boxes); these are partially or fully base-paired in the monomer but become unpaired as a result of the RNA rearrangements accompanying the intermolecular base pairing of PAL1 and PAL2. These bases are a crucial element in ψ, as replacement of the four UCUG sequences with UCUA prevents selective packaging of the viral RNA; the exposure of these bases in dimers, but not monomers, presumably explains the selective packaging of dimeric RNA [20]. (Figure reproduced from Trends in Biochemical Sciences, Copyright 2011, with permission from Elsevier [21].)
