Abstract
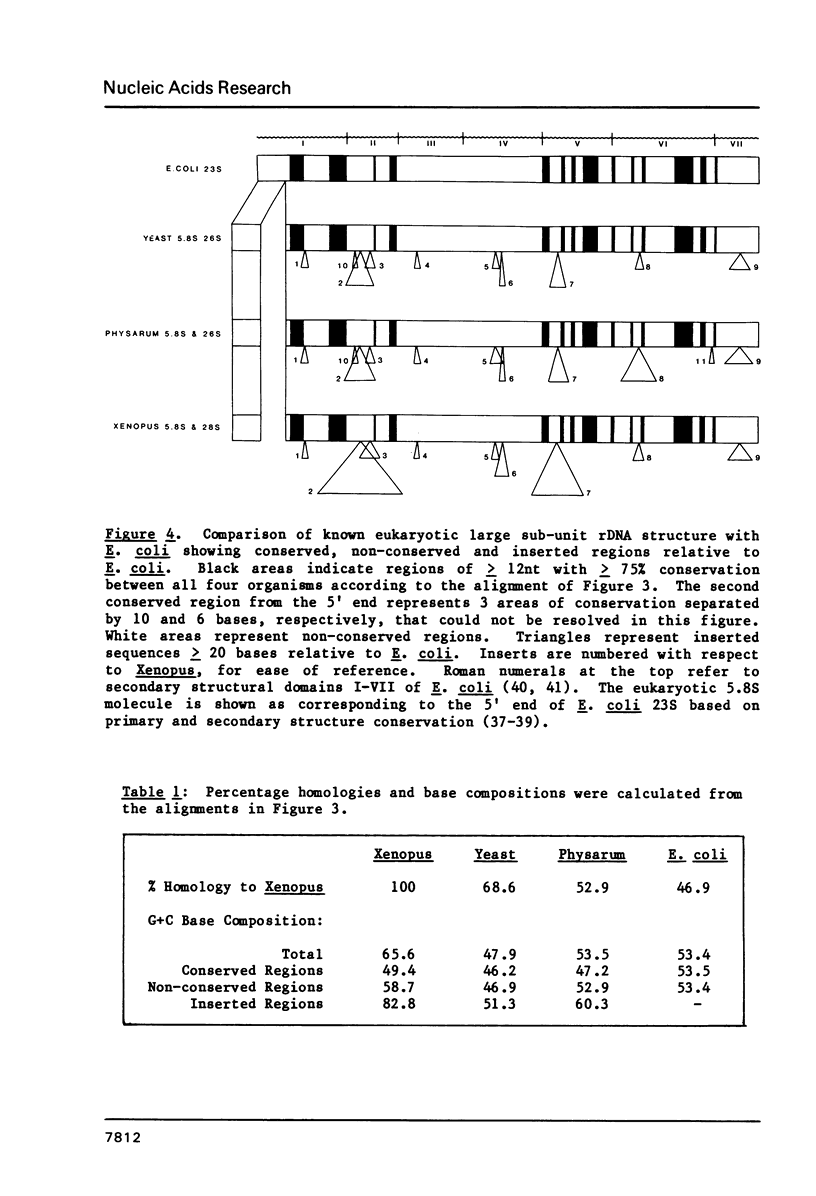
We have determined the complete nucleotide sequence of Xenopus laevis 28S rDNA (4110 bp). In order to locate evolutionarily conserved regions within rDNA, we compared the Xenopus 28S sequence to homologous rDNA sequences from yeast, Physarum, and E. coli. Numerous regions of sequence homology are dispersed throughout the entire length of rDNA from all four organisms. These conserved regions have a higher A + T base composition than the remainder of the rDNA. The Xenopus 28S rDNA has nine major areas of sequence inserted when compared to E. coli 23S rDNA. The total base composition of these inserts in Xenopus is 83% G + C, and is generally responsible for the high (66%) G + C content of Xenopus 28S rDNA as a whole. Although the length of the inserted sequences varies, the inserts are found in the same relative positions in yeast 26S, Physarum 26S, and Xenopus 28S rDNAs. In one insert there are 25 bases completely conserved between the various eukaryotes, suggesting that this area is important for eukaryotic ribosomes. The other inserts differ in sequence between species and may or may not play a functional role.
Full text
PDF




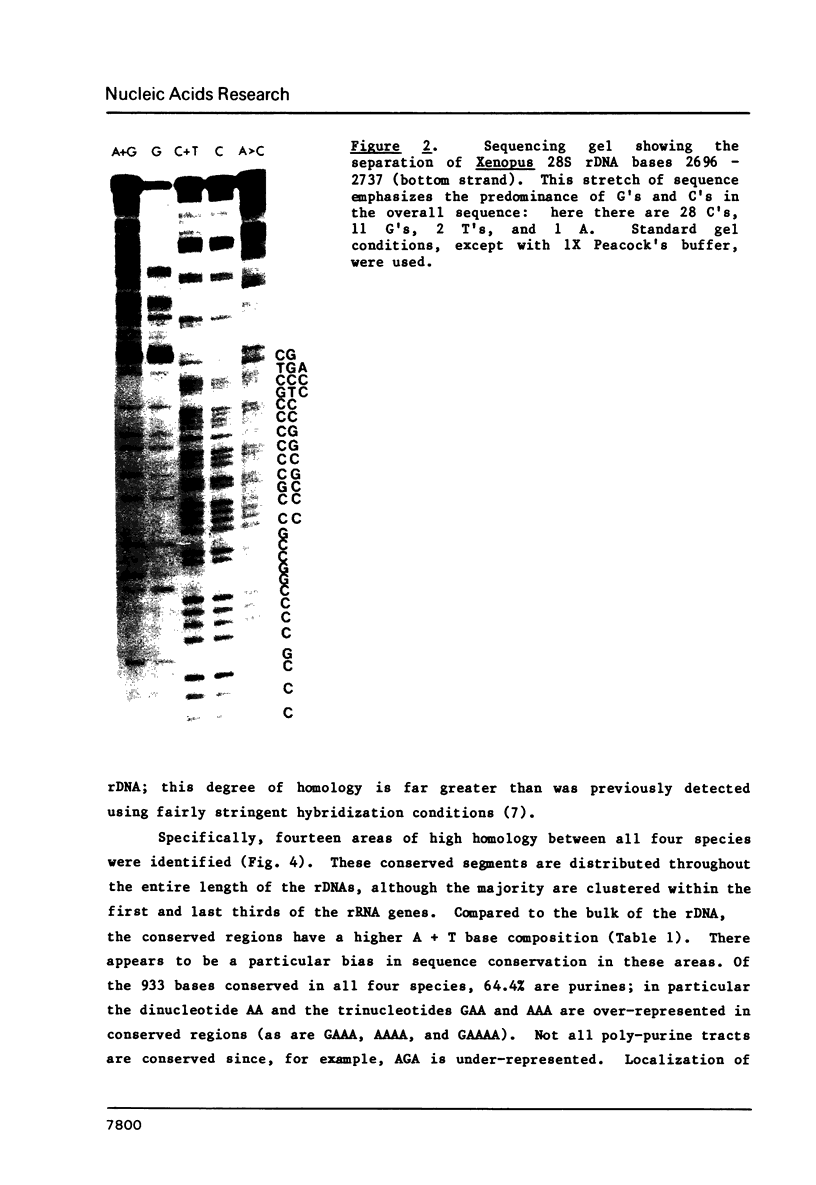
















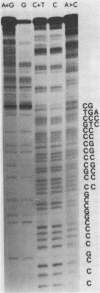
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amaldi F. Non-random variability in evolution of base compositions of ribosomal RNA. Nature. 1969 Jan 4;221(5175):95–96. doi: 10.1038/221095a0. [DOI] [PubMed] [Google Scholar]
- Birnstiel M., Speirs J., Purdom I., Jones K., Loening U. E. Properties and composition of the isolated ribosomal DNA satellite of Xenopus laevis. Nature. 1968 Aug 3;219(5153):454–463. doi: 10.1038/219454a0. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boseley P., Moss T., Mächler M., Portmann R., Birnstiel M. Sequence organization of the spacer DNA in a ribosomal gene unit of Xenopus laevis. Cell. 1979 May;17(1):19–31. doi: 10.1016/0092-8674(79)90291-5. [DOI] [PubMed] [Google Scholar]
- Brand R. C., Gerbi S. A. Fine structure of ribosomal RNA. II. Distribution of methylated sequences within Xenopus laevis rRNA. Nucleic Acids Res. 1979 Nov 24;7(6):1497–1511. doi: 10.1093/nar/7.6.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branlant C., Krol A., Machatt M. A., Pouyet J., Ebel J. P., Edwards K., Kössel H. Primary and secondary structures of Escherichia coli MRE 600 23S ribosomal RNA. Comparison with models of secondary structure for maize chloroplast 23S rRNA and for large portions of mouse and human 16S mitochondrial rRNAs. Nucleic Acids Res. 1981 Sep 11;9(17):4303–4324. doi: 10.1093/nar/9.17.4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Cartwright E., McShane T., Williamson R. The nucleotide sequence of somatic 5 S RNA from Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):8–12. doi: 10.1016/0014-5793(72)80442-3. [DOI] [PubMed] [Google Scholar]
- Clark C. G., Gerbi S. A. Ribosomal RNA evolution by fragmentation of the 23S progenitor: maturation pathway parallels evolutionary emergence. J Mol Evol. 1982;18(5):329–336. doi: 10.1007/BF01733899. [DOI] [PubMed] [Google Scholar]
- Cox R. A., Thompson R. D. Distribution of sequences common to the 25--28S-ribonucleic acid genes of Xenopus laevis and Neurospora crassa. Biochem J. 1980 Apr 1;187(1):75–90. doi: 10.1042/bj1870075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick F. H. The origin of the genetic code. J Mol Biol. 1968 Dec;38(3):367–379. doi: 10.1016/0022-2836(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Dawid I. B., Wellauer P. K. A reinvestigation of 5' leads to 3' polarity in 40S ribosomal RNA precursor of Xenopus laevis. Cell. 1976 Jul;8(3):443–448. doi: 10.1016/0092-8674(76)90157-4. [DOI] [PubMed] [Google Scholar]
- Ford P. J., Southern E. M. Different sequences for 5S RNA in kidney cells and ovaries of Xenopus laevis. Nat New Biol. 1973 Jan 3;241(105):7–12. doi: 10.1038/newbio241007a0. [DOI] [PubMed] [Google Scholar]
- Frank R., Müller D., Wolff C. Identification and suppression of secondary structures formed from deoxy-oligonucleotides during electrophoresis in denaturing polyacrylamide-gels. Nucleic Acids Res. 1981 Oct 10;9(19):4967–4979. doi: 10.1093/nar/9.19.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Georgiev O. I., Nikolaev N., Hadjiolov A. A., Skryabin K. G., Zakharyev V. M., Bayev A. A. The structure of the yeast ribosomal RNA genes. 4. Complete sequence of the 25 S rRNA gene from Saccharomyces cerevisae. Nucleic Acids Res. 1981 Dec 21;9(24):6953–6958. doi: 10.1093/nar/9.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbi S. A. Fine structure of ribosomal RNA. I. Conservation of homologous regions within ribosomal RNA of eukaryotes. J Mol Biol. 1976 Sep 25;106(3):791–816. doi: 10.1016/0022-2836(76)90265-5. [DOI] [PubMed] [Google Scholar]
- Glotz C., Zwieb C., Brimacombe R., Edwards K., Kössel H. Secondary structure of the large subunit ribosomal RNA from Escherichia coli, Zea mays chloroplast, and human and mouse mitochondrial ribosomes. Nucleic Acids Res. 1981 Jul 24;9(14):3287–3306. doi: 10.1093/nar/9.14.3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. III. Location of evolutionarily conserved regions within ribosomal DNA. J Mol Biol. 1980 Jun 25;140(2):321–339. doi: 10.1016/0022-2836(80)90109-6. [DOI] [PubMed] [Google Scholar]
- Gourse R. L., Gerbi S. A. Fine structure of ribosomal RNA. IV. Extraordinary evolutionary conservation in sequences that flank introns in rDNA. Nucleic Acids Res. 1980 Aug 25;8(16):3623–3637. doi: 10.1093/nar/8.16.3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourse R. L., Thurlow D. L., Gerbi S. A., Zimmermann R. A. Specific binding of a prokaryotic ribosomal protein to a eukaryotic ribosomal RNA: implications for evolution and autoregulation. Proc Natl Acad Sci U S A. 1981 May;78(5):2722–2726. doi: 10.1073/pnas.78.5.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofstetter H., Kressman A., Birnstiel M. L. A split promoter for a eucaryotic tRNA gene. Cell. 1981 May;24(2):573–585. doi: 10.1016/0092-8674(81)90348-2. [DOI] [PubMed] [Google Scholar]
- Jacq B. Sequence homologies between eukaryotic 5.8S rRNA and the 5' end of prokaryotic 23S rRNa: evidences for a common evolutionary origin. Nucleic Acids Res. 1981 Jun 25;9(12):2913–2932. doi: 10.1093/nar/9.12.2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kressmann A., Hofstetter H., Di Capua E., Grosschedl R., Birnstiel M. L. A tRNA gene of Xenopus laevis contains at least two sites promoting transcription. Nucleic Acids Res. 1979 Dec 11;7(7):1749–1763. doi: 10.1093/nar/7.7.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lava-Sanchez P. A., Amaldi F., Posta A. L. Base composition of ribosomal RNA and evolution. J Mol Evol. 1972 Dec 29;2(1):44–55. doi: 10.1007/BF01653942. [DOI] [PubMed] [Google Scholar]
- Loening U. E., Jones K. W., Birnstiel M. L. Properties of the ribosomal RNA precursor in Xenopus laevis; comparison to the precursor in mammals and in plants. J Mol Biol. 1969 Oct 28;45(2):353–366. doi: 10.1016/0022-2836(69)90110-7. [DOI] [PubMed] [Google Scholar]
- Machatt M. A., Ebel J. P., Branlant C. The 3'-terminal region of bacterial 23S ribosomal RNA: structure and homology with the 3'-terminal region of eukaryotic 28S rRNA and with chloroplast 4.5s rRNA. Nucleic Acids Res. 1981 Apr 10;9(7):1533–1549. doi: 10.1093/nar/9.7.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow J. F., Cohen S. N., Chang A. C., Boyer H. W., Goodman H. M., Helling R. B. Replication and transcription of eukaryotic DNA in Escherichia coli. Proc Natl Acad Sci U S A. 1974 May;71(5):1743–1747. doi: 10.1073/pnas.71.5.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss T., Boseley P. G., Birnstiel M. L. More ribosomal spacer sequences from Xenopus laevis. Nucleic Acids Res. 1980 Feb 11;8(3):467–485. doi: 10.1093/nar/8.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F., Clarkson S. G. Nucleotide sequence of genes coding for tRNAPhe and tRNATyr from a repeating unit of X. laevis DNA. Cell. 1980 Feb;19(2):345–353. doi: 10.1016/0092-8674(80)90509-7. [DOI] [PubMed] [Google Scholar]
- Nazar R. N. A 5.8 S rRNA-like sequence in prokaryotic 23 S rRNA. FEBS Lett. 1980 Oct 6;119(2):212–214. doi: 10.1016/0014-5793(80)80254-7. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O. Role of the 5'-terminal sequence in the RNA binding site of yeast 5.8 S rRNA. FEBS Lett. 1980 Jun 16;115(1):71–76. doi: 10.1016/0014-5793(80)80729-0. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Kop J., Wheaton V., Brosius J., Gutell R. R., Kopylov A. M., Dohme F., Herr W., Stahl D. A., Gupta R. Secondary structure model for 23S ribosomal RNA. Nucleic Acids Res. 1981 Nov 25;9(22):6167–6189. doi: 10.1093/nar/9.22.6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orgel L. E. Evolution of the genetic apparatus. J Mol Biol. 1968 Dec;38(3):381–393. doi: 10.1016/0022-2836(68)90393-8. [DOI] [PubMed] [Google Scholar]
- Otsuka T., Nomiyama H., Yoshida H., Kukita T., Kuhara S., Sakaki Y. Complete nucleotide sequence of the 26S rRNA gene of Physarum polycephalum: its significance in gene evolution. Proc Natl Acad Sci U S A. 1983 Jun;80(11):3163–3167. doi: 10.1073/pnas.80.11.3163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Peacock A. C., Dingman C. W. Molecular weight estimation and separation of ribonucleic acid by electrophoresis in agarose-acrylamide composite gels. Biochemistry. 1968 Feb;7(2):668–674. doi: 10.1021/bi00842a023. [DOI] [PubMed] [Google Scholar]
- Queen C. L., Korn L. J. Computer analysis of nucleic acids and proteins. Methods Enzymol. 1980;65(1):595–609. doi: 10.1016/s0076-6879(80)65062-9. [DOI] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence encoding the 5' end of Xenopus laevis 18S rRNA. Nucleic Acids Res. 1980 Jul 11;8(13):2871–2884. doi: 10.1093/nar/8.13.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim M., Maden B. E. Nucleotide sequence of Xenopus laevis 18S ribosomal RNA inferred from gene sequence. Nature. 1981 May 21;291(5812):205–208. doi: 10.1038/291205a0. [DOI] [PubMed] [Google Scholar]
- Sege R., Söll D., Ruddle F. H., Queen C. A conversational system for the computer analysis of nucleic acid sequences. Nucleic Acids Res. 1981 Jan 24;9(2):437–444. doi: 10.1093/nar/9.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seif I., Khoury G., Dhar R. A rapid enzymatic DNA sequencing technique: determination of sequence alterations in early simian virus 40 temperature sensitive and deletion mutants. Nucleic Acids Res. 1980 May 24;8(10):2225–2240. doi: 10.1093/nar/8.10.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair J. H., Brown D. D. Retention of common nucleotide sequences in the ribosomal deoxyribonucleic acid of eukaryotes and some of their physical characteristics. Biochemistry. 1971 Jul 6;10(14):2761–2769. doi: 10.1021/bi00790a017. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Birnstiel M. L. A simple method for DNA restriction site mapping. Nucleic Acids Res. 1976 Sep;3(9):2387–2398. doi: 10.1093/nar/3.9.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Regt V. C., Planta R. J., Branlant C., Krol A., Ebel J. P. The primary and secondary structure of yeast 26S rRNA. Nucleic Acids Res. 1981 Dec 21;9(24):6935–6952. doi: 10.1093/nar/9.24.6935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegnez M., Monier R., Denis H. Sequence heterogeneity of 5 S RNA in Xenopus laevis. FEBS Lett. 1972 Sep 1;25(1):13–20. doi: 10.1016/0014-5793(72)80443-5. [DOI] [PubMed] [Google Scholar]