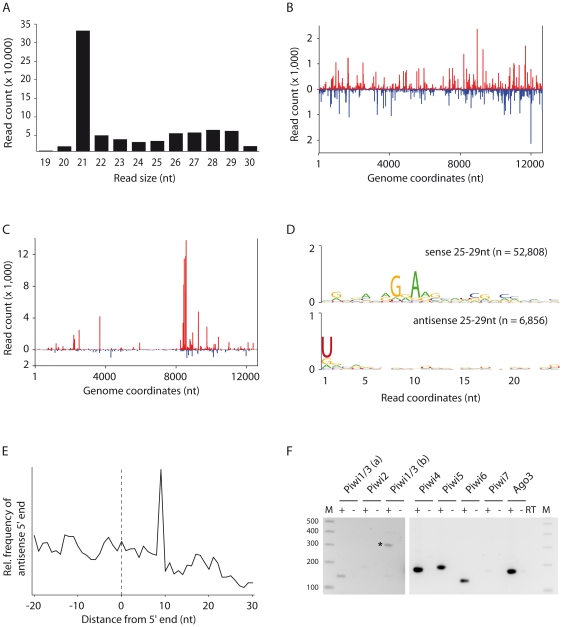
Figure 4. Aedes aegypti Aag2 cells produce vsiRNA and vpiRNA with a ping-pong signature upon arbovirus infection.
A. Size distribution of the small RNA reads that match the genome of SINV-GFP with 0 mismatches. Profile of 21 nt vsiRNAs (B) and 25–29 nt (C) SINV-GFP-derived small RNAs allowing 0 mismatch during alignment. Viral small RNA that mapped to the sense and antisense strand of the SINV-GFP genome are shown in red and blue, respectively. D. Conservation and relative nucleotide frequency per position of 25–29 nt SINV-GFP-derived reads that mapped to the sense (top) and antisense (bottom) strands of the SINV-GFP genome. n indicates the number of reads used to generate each logo. E. Frequency map of the distance between 25–29 nt small RNAs that mapped to opposite strands of the SINV-GFP genome. The peak at position 9 on the sequence (the first nucleotide being position 0) indicates the position of maximal probability of finding the 5′ end of a complementary small RNA. F. Expression of PIWI family members in Aag2 cells analyzed by RT-PCR. cDNA synthesis was performed in the presence (+) or absence (−) of reverse transcriptase (RT). The -RT samples are included as controls for contamination of RNA preparations with chromosomal DNA. The coding sequences of Piwi1 and Piwi3 are 95% identical at the nucleotide level. Two different primer sets that amplify both Piwi1 and Piwi3 were used (a and b). A higher exposure was used for the gel image with Piwi1to Piwi3. A 100 bp ladder was used as a size marker (M). The asterisk indicates a non-specific PCR amplification product.