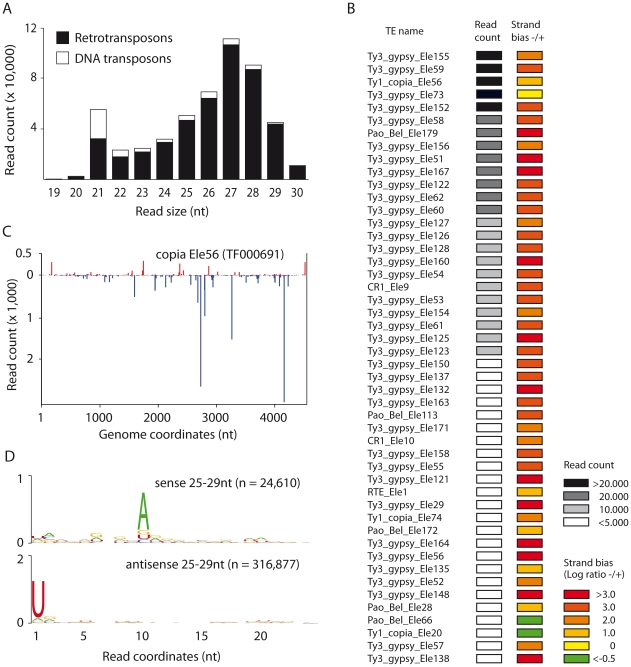
Figure 5. Aedes aegypti Aag2 cells produce transposon-derived piRNAs with a ping-pong signature.
A. Size distribution of the small RNA reads that match with 0 mismatches against an Aedes aegypti transposon dataset that contain full-length non-composite transposons sequences (TEfam: http://tefam.biochem.vt.edu/tefam/index.php). B. Heat map for 25–29 nt small RNAs that mapped to individual retrotransposons with more than 1000 reads. Read count and log-transformed ratios of antisense/sense small RNAs are presented. C. Profile of 25–29 nt reads that mapped to the transposon Copia Ele56 (TF000691) allowing 0 mismatch during alignment. Transposon-derived piRNAs that mapped to the sense and antisense strand of the transposon sequence are shown in red and blue, respectively. D. Conservation and relative nucleotide frequency per position of 25–29 nt reads that mapped to the sense (top) and the antisense (bottom) strands of the entire transposon dataset. n indicates the number of reads used to generate each logo.