Abstract
Flow cytometric analysis is a reliable and convenient method for investigating molecules at the single cell level. Previously, recombinant human immunodeficiency virus type 1 (HIV-1) strains were constructed that express a fluorescent reporter, either enhanced green fluorescent protein, or DsRed, which allow the monitoring of HIV-1-infected cells by flow cytometry. The present study further investigated the potential of these recombinant viruses in terms of whether the HIV-1 fluorescent reporters would be helpful in evaluating viral replication based on fluorescence intensity. When primary CD4+ T cells were infected with recombinant viruses, the fluorescent reporter intensity measured by flow cytometry was associated with the level of CD4 downmodulation and Gag p24 expression in infected cells. Interestingly, some HIV-1-infected cells, in which CD4 was only moderately downmodulated, were reporter-positive but Gag p24-negative. Furthermore, when the activation status of primary CD4+ T cells was modulated by T cell receptor-mediated stimulation, we confirmed the preferential viral production upon strong stimulation and showed that the intensity of the fluorescent reporter within a proportion of HIV-1-infected cells was correlated with the viral replication level. These findings indicate that a fluorescent reporter encoded within HIV-1 is useful for the sensitive detection of productively infected cells at different stages of infection and for evaluating cell-associated viral replication at the single cell level.
Keywords: HIV-1, flow cytometry, EGFP, DsRed, Gag, productive infection
Introduction
Human immunodeficiency virus type 1 (HIV-1) interacts with its primary receptor, CD4, and a co-receptor, usually CCR5 or CXCR4, to infect T cells, macrophages, and dendritic cells (Mcclure et al., 1987; Berger et al., 1999; Tsunetsugu-Yokota, 2008). Single cell analysis of HIV-1-infected cells is an essential approach to investigate the differential dynamics of HIV-1 infection and the cellular consequences for each of the HIV-1-targeted cell populations. To monitor HIV-1 infection, a recombinant HIV-1 encoding a reporter luciferase (Luc) gene, or indicator cells transduced with enzymatic reporters such as Luc, β-galactosidase, alkaline phosphatase, and chloramphenicol acetyl transferase, incorporated downstream of the HIV-1 long terminal repeats (LTR) have been widely used (Kar-Roy et al., 2000). However, these reporters require additional substrates or co-factors, and lysis or fixation of cells is required to show reporter activity, which makes the experimental process more complex. In addition, it is difficult to distinguish infected cells from uninfected cells using these reporter assays.
An alternative molecule, green fluorescent protein (GFP) and/or its derivatives, is a powerful reporter that does not require any substrates and co-factors to generate a reporter signal (Chalfie, 1995; Cubitt et al., 1995; Heim et al., 1995). Page et al. (1997) first used a GFP derivative, called enhanced green fluorescent protein (EGFP), as a fluorescent reporter molecule for HIV-1 and showed that infected cells were detectable and, more importantly, distinguishable from uninfected cells using flow cytometry. Furthermore, a red fluorescent protein, DsRed, has been used as an HIV-1 fluorescent reporter (Weber et al., 2006). The main benefit of such recombinant HIV-1 molecules is that the targeted cells do not require any modulation (e.g., transfection) of exogenous reporter genes and, therefore, they allow the characterization of intact HIV-1-infected cells. In most cases of previous recombinant HIV-1 strains, the nef gene was replaced with a reporter gene. Therefore, we previously constructed nef-intact, replication-competent, recombinant HIV-1 strains encoding either EGFP or DsRed, and showed that CXCR4-tropic X4 and CCR5-tropic R5 viruses replicate differently in CD4+ T cells simultaneously infected with X4 HIV-1 encoding EGFP and R5 HIV-1 encoding DsRed (Yamamoto et al., 2009). Such recombinant HIV-1 strains encoding a fluorescent reporter gene will be even more valuable because of recent advances in multicolor flow cytometry, which permit more detailed characterization of HIV-1-infected cells.
Flow cytometry is a reliable and convenient method for analysis at the single cell level. Because the transcriptional activity of HIV-1 can be quantitatively monitored in indicator cells according to the fluorescence intensity of an EGFP reporter driven by the HIV-1 LTR (Dorsky et al., 1996; Gervaix et al., 1997; Kar-Roy et al., 2000), we investigated whether the HIV-1-expressing fluorescent reporters EGFP and DsRed would allow the quantitative evaluation of viral replication using a flow cytometer. The results show that a fluorescent reporter signal generated by recombinant HIV-1 strains enables the detection of infected cells at various stages of the viral life cycle.
Materials and Methods
Cell preparation
Human peripheral blood samples were collected from healthy donors after written informed consent. Sample collection was approved by the Institutional Ethical Committee of the National Institute of Infectious Diseases (NIID; Tokyo, Japan). Peripheral blood mononuclear cells (PBMCs) were separated on a Ficoll–Hypaque density gradient (Lymphosepal; IBL, Gunma, Japan) and CD4+ T cells were negatively selected from the PBMCs using an EasySep Human CD4+ T cell Enrichment Kit (StemCell Technologies, Vancouver, BC, Canada).
CEM cells stably expressing human CCR5 (CEM–CCR5) were established by transducing CEM cells with the human ccr5 gene using a conventional mouse retrovirus system. CEM–CCR5 cells were maintained in complete RPMI medium (10% heat-inactivated fetal bovine serum, 100 μg/ml penicillin, 100 μg/ml streptomycin, and 2 mM l-glutamine) supplemented with 1 μg/ml puromycin at 37°C.
Preparation of HIV-1 virus stocks
We previously constructed pNL432-based proviral clones encoding EGFP (pNL-E) or DsRed (pNL-D) for X4-tropic HIV-1NL-E or HIV-1NL-D, respectively, and pNLAD8-based proviral clones encoding EGFP (pNLAD8-E) or DsRed (pNLAD8-D) for R5-tropic HIV-1NLAD8-E or HIV-1NLAD8-D, respectively (Yamamoto et al., 2009; Figure 1). To prepare the HIV-1 viral stocks, the human embryonic kidney cell line 293T was transfected with pNL-E, pNL-D, pNLAD8-E, or pNLAD8-D using the calcium phosphate precipitation method and then incubated for 48 h. Culture supernatants were filtered and frozen at −80°C. The amount of virus in each culture supernatant was measured using an in-house HIV-1 Gag p24 enzyme-linked immunosorbent assay (ELISA; Tsunetsugu-Yokota et al., 1995).
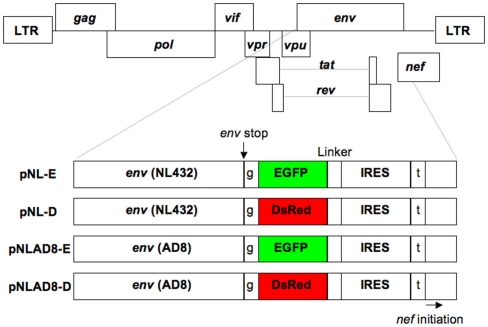
Figure 1.
Structure of the proviral DNA. The pNL432-based proviral clones encoded EGFP (pNL-E) or DsRed (pNL-D) for X4-tropic HIV-1NL-E or HIV-1NL-D, respectively, and the pNLAD8-based proviral clones encoded EGFP (pNLAD8-E) or DsRed (pNLAD8-D) for R5-tropic HIV-1NLAD8-E or HIV-1NLAD8-D, respectively. EGFP or DsRed was not expressed as a fusion protein with Env due to the insertion of a single base after the Env stop codon. Nef was also independently expressed under the control of IRES. t (Thymine) and g (guanine) are additional DNA sequences.
Stimulation of T cell receptors
T cell receptors (TCR) were stimulated as described previously (Yamamoto et al., 2009) with some modifications. In brief, primary CD4+ T cells were suspended in complete RPMI medium supplemented with 5% human plasma and stimulated with 5 μg/ml of immobilized anti-human CD3 monoclonal antibody (mAb; eBioscience, San Diego, CA) and 1 μg/ml of soluble anti-human CD28 mAb (eBioscience) in U-bottom, 96-well plates at 37°C for 4 (weak stimulation) or 24 h (strong stimulation).
HIV-1 infection and cell culture
Primary CD4+ T cells (either unstimulated or pre-TCR-stimulated) or CEM–CCR5 cells were infected with 200 ng of p24-measured amounts of HIV-1NL-E, HIV-1NL-D, HIV-1NLAD8-E, or HIV-1NLAD8-D per 1 × 106 cells by spinoculation at 1200 × g for 2 h at 25 (conventional conditions) or 4°C (for CEM–CCR5 cells), as described previously (O’doherty et al., 2000; Dai et al., 2009). After spinoculation, cells were washed three times with PBS. Primary CD4+ T cells were then suspended in complete RPMI medium supplemented with 5% human plasma. The cell suspensions derived from unstimulated or pre-TCR-stimulated CD4+ T cells were settled onto U-bottom, 96-well plates with or without TCR-stimulation, respectively, at 37°C for 24 h. After the 24 h culture, cells were washed three times with PBS, suspended in complete RPMI medium supplemented with 5% human plasma and 50 U/ml recombinant interleukin-2, and cultured in U-bottom, 96-well plates at 37°C for up to 4 days.
Flow cytometry
Cells were stained with fluorescence-conjugated mAbs as described previously (Yamamoto et al., 2009). The following mAbs were used for flow cytometry in various combinations: Pacific Blue-conjugated anti-human CD3 mAb (BioLegend, San Diego, CA, USA), phycoerythrin Cy7-conjugated anti-human CD4 mAb (BioLegend), and Alexa Fluor 700-conjugated anti-human CD8a mAb (BioLegend); and Nu24 mAb specific for HIV-1 Gag p24 (kindly provided by Dr. T. Sata, NIID, Tokyo, Japan) and conjugated to Alexa Fluor 647 using an Alexa Fluor 647 Protein Labeling Kit (Molecular Probes, Eugene, OR, USA). Dead cells were stained with propidium iodide or a LIVE/DEAD Fixable Dead Cell Stain Kit (L34957; Invitrogen, Carlsbad, CA, USA). Intracellular staining (ICS) by Nu24 mAb was performed using a FIX and PERM Fixation and Permeabilization Kit (Invitrogen). Data collection was performed using a FACSCanto II (BD Bioscience, San Diego, CA, USA) and the data were analyzed using FACSDiva software (BD Bioscience) and FlowJo software (Tree Star, San Carlos, CA, USA).
Quantification of replicated HIV-1 in cell culture supernatants
Human immunodeficiency virus type 1 replication was quantified in cell culture supernatants by ELISA and real-time RT-PCR. Gag p24 was measured using a RETRO-TEK HIV-1 p24 Antigen ELISA (ZeptoMetrix Corporation, Buffalo, NY, USA). For real-time RT-PCR, viral RNA was extracted using a QIAamp Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) and subjected to real-time RT-PCR using a SuperScript III Platinum One-Step Quantitative RT-PCR System (Invitrogen), a set of HIV-1 gag-targeted primers, and a TaqMan probe as previously described (Saito et al., 2010). PCR was performed in an Mx3000P (Stratagene, La Jolla, CA, USA).
Results
CD4 downmodulation is associated with HIV-1 fluorescent reporter intensity
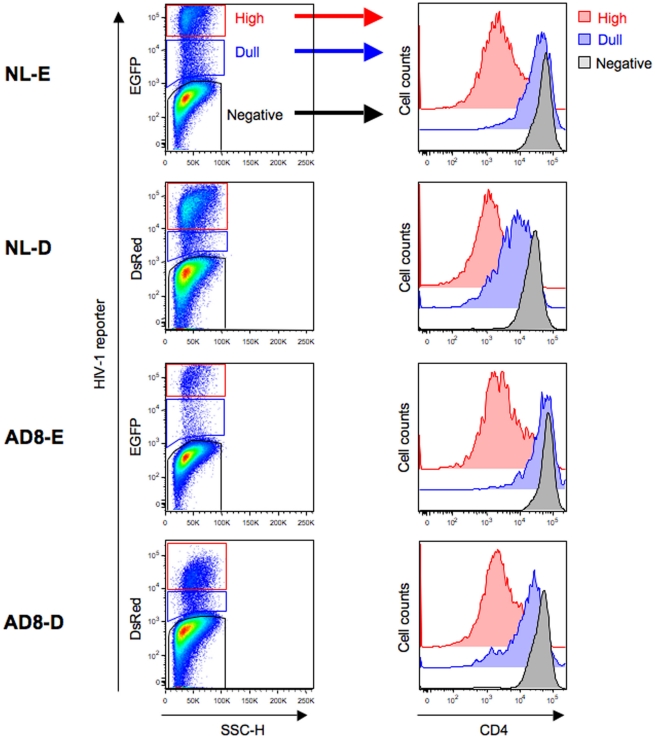
The cell surface CD4 molecule is downmodulated in HIV-1-infected cells in response to the HIV-1 components Env, Nef, and Vpu (Malim and Emerman, 2008). Therefore, to investigate the correlation between the level of CD4 downmodulation and the HIV-1 fluorescent reporter intensity, primary CD4+ T cells infected with HIV-1NL-E, HIV-1NL-D, HIV-1NLAD8-E, or HIV-1NLAD8-D followed by TCR-stimulation for 1 day and cultivation for a further 4 days were analyzed by flow cytometry. As shown in Figure 2 (left panels), HIV-1-infected cells expressing a fluorescent reporter signal, EGFP, or DsRed, were detected, although the numbers varied between individual donors (n = 3–4): about 10–30% for X4-tropic HIV-1NL-E-infected and HIV-1NL-D-infected cells and 1–10% for R5-tropic HIV-1NLAD8-E-infected and HIV-1NLAD8-D-infected cells. However, the number of HIV-1+ cells was comparable between HIV-1NL-E and HIV-1NL-D (X4-tropic), and between HIV-1NLAD8-E and HIV-1NLAD8-D (R5-tropic) within each donor, showing that the fluorescent reporter genes encoded within the HIV-1 proviral genome did not affect HIV-1 infectivity as described previously (Yamamoto et al., 2009). When we categorized CD3+CD8− T cells into three fractions (HIV-1-negative, -dull, and -high) based on the fluorescence intensity of EGFP and DsRed, we found that CD4 was strongly downmodulated in the HIV-1 high fraction in all the HIV-1 strains (Figure 2, right panels). Interestingly, CD4 was also downmodulated in the HIV-1 dull fraction, but the level was modest compared with that in the HIV-1 high fraction (Figure 2, right panels). These results indicate that the level of CD4 downmodulation is associated with HIV-1 fluorescent reporter intensity.
Figure 2.
Flow cytometry analysis of HIV-1-infected cells. Unstimulated CD4+ T cells were infected with HIV-1NL-E, HIV-1NL-D, HIV-1NLAD8-E, or HIV-1NLAD8-D and cultured for 5 days (including the initial 1 day culture with TCR-stimulation). (Left panels) representative pseudo-color plot profiles for the Dead−/CD3+/CD8−-gated cell fractions from three donors. Cells were classified as high (red), dull (blue), and negative (black) based on the HIV-1 reporter intensity. (Right panels) histograms of CD4 expression by the high (red), dull (blue), and negative (black) fractions defined.
Fixation/permeabilization weakens the HIV-1 fluorescent reporter signal
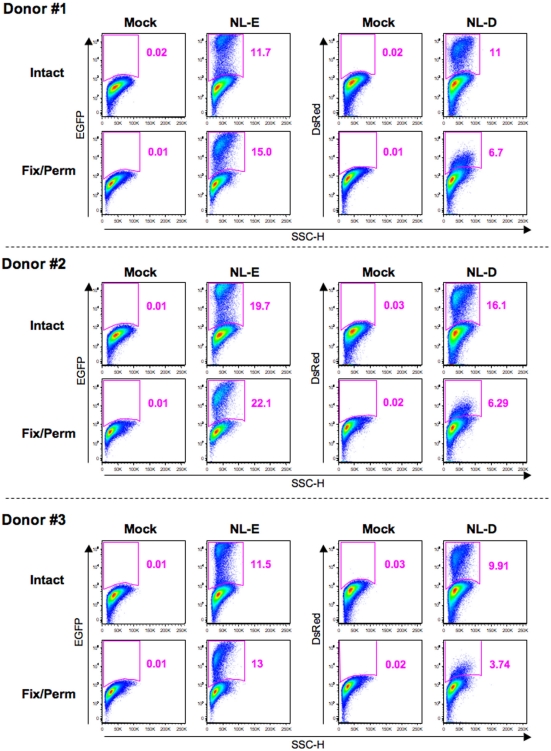
To investigate the correlation between HIV-1 fluorescent reporter intensity and viral replication levels, we attempted to perform ICS for Gag p24 in HIV-1-infected cells prepared as described above. When we observed X4-tropic HIV-1NL-E-infected and HIV-1NL-D-infected cells from three donors by flow cytometry, we noticed that fixation/permeabilization, an essential step for ICS, weakened the fluorescent reporter signal for both EGFP and DsRed. Figure 3 shows the flow cytometry profiles obtained for EGFP and DsRed at identical photomultiplier tube (PMT) voltages between intact (untreated) cells and fixed/permeabilized cells to visualize the differences in fluorescent reporter intensity. DsRed+ cells were not properly separated from DsRed− cells within the population treated by fixation/permeabilization; the frequency of DsRed+ cells was, therefore, markedly decreased. No adjustment of the flow cytometer settings, including PMT voltage and compensation, improved the blunted fluorescent reporter signal generated after fixation/permeabilization. Nevertheless, the number of EGFP+ cells within the intact cell and fixed/permeabilized cell populations was comparable. Similar results were obtained for R5-tropic HIV-1NLAD8-E and HIV-1NLAD8-D (data not shown). Taken together, these results indicate that it is preferable to use an EGFP reporter when the fixation/permeabilization of cells is required.
Figure 3.
Influence of fixation/permeabilization treatment on HIV-1 fluorescent reporter signals. Pseudo-color plot profiles for the Dead−/CD3+/CD8−-gated cell fractions from the mock, HIV-1NL-E infection (NL-E), and HIV-1NL-D infection (NL-D) groups from all three donors tested. Analyzed cells were prepared as outlined in the legend to Figure 2 and then either fixed/permeabilized (fix/perm) or not (intact).
HIV-1 fluorescent reporter signals reliably detect productively infected cells showing different viral replication levels
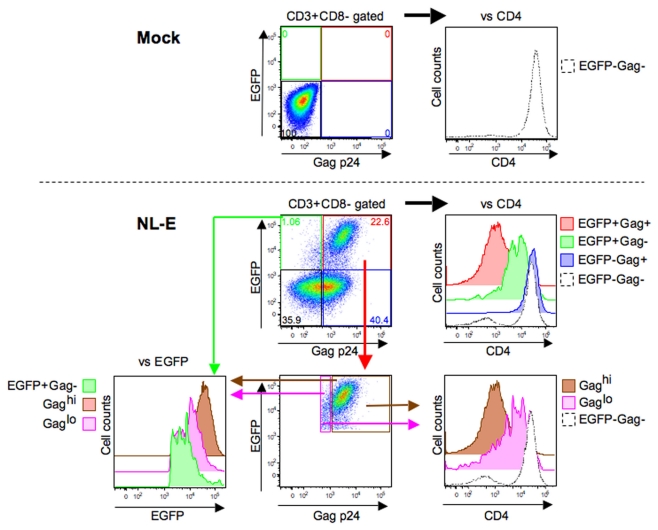
Following the results shown in Figure 3, we next assessed viral replication levels in the HIV-1NL-E infection group (5 days culture) from six donors using Gag p24 ICS (Figure 4). A representative flow cytometric analysis showed that not all EGFP+ cells were Gag+ and vice versa. When CD4 expression levels were compared in each of the four cell fractions based on the expression patterns of EGFP and Gag p24 (EGFP+Gag+, EGFP+Gag−, EGFP−Gag+, and EGFP−Gag−), the strongest downmodulation of CD4 was observed in EGFP+Gag+ cells (red fraction). CD4 downmodulation was moderate in EGFP+Gag− cells (green fraction). However, CD4 was not downmodulated at all in EGFP−Gag+ cells (blue fraction) and the expression level of CD4 was the same as that in EGFP−Gag− cells (black fraction). We further divided the EGFP+Gag+ cells (red fraction) into Gaghi (brown fraction) and Gaglo cells (pink fraction) and compared the expression levels of EGFP and CD4 with those of Gag p24. Gaghi cells (brown fraction) showed the strongest expression of EGFP and the strongest downmodulation of CD4. Gaglo cells (pink fraction) showed an intermediate level of EGFP expression [between that of Gaghi cells (brown fraction) and that of EGFP+Gag− cells (green fraction)] and CD4 expression [between that of Gaghi cells (brown fraction) and EGFP−Gag− cells (black fraction)]. These results indicate that the expression level of EGFP correlates with that of Gag p24 in HIV-1-infected cells in which CD4 is downmodulated.
Figure 4.
Correlation between HIV-1 fluorescent reporter signal and Gag p24 expression. Representative flow cytometric analyses of the mock and HIV-1NL-E infection groups from six donors. These infection groups were prepared as outlined in the legend to Figure 2. The EGFP+Gag+ (red), EGFP+Gag− (green), EGFP−Gag+ (blue), EGFP−Gag− (black), Gaghi (brown), and Gaglo (pink) cell fractions were categorized based on the expression patterns of EGFP and Gag p24. The expression levels of CD4 and EGFP in each cell fraction were compared according to their histogram profiles.
HIV-1-bound or -internalized cells are also detected by Gag p24 ICS
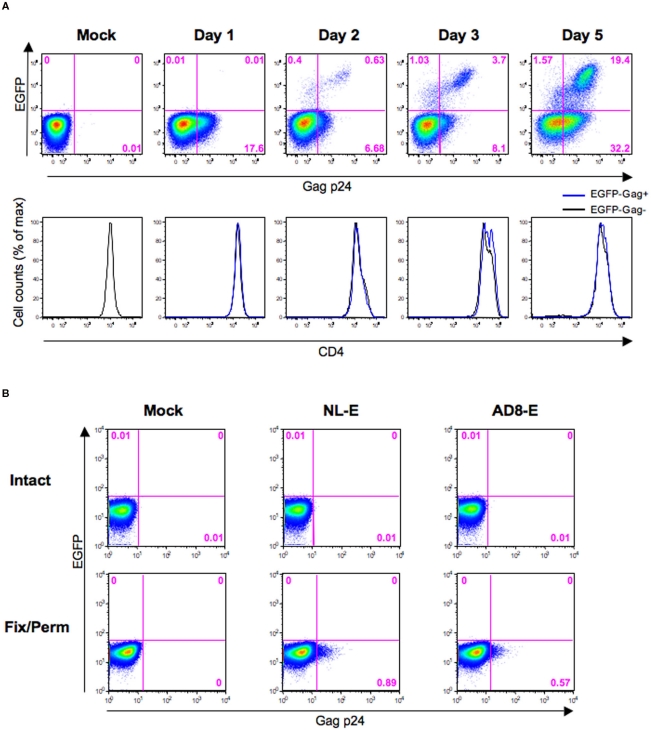
Because CD4 downmodulation was not observed in EGFP−Gag+ cells (Figure 4; blue fraction), it is possible that these cells may still be bound by or have internalized HIV-1 but have not produced virions. Therefore, we next investigated the kinetics of EGFP−Gag+ cells during 5 days post-infection. Primary CD4+ T cells from three donors were infected with HIV-1NL-E followed by TCR-stimulation for 1 day and cultivation for a further 4 days. Figure 5A shows a representative flow cytometric analysis. At 1 day post-infection, 17.6% of Gag p24+ cells were observed, despite the fact that no EGFP+ cells were detected. At 2 days post-infection, the proportion of EGFP−Gag+ cells was decreased and EGFP+ cells including Gag p24+ and Gag p24− cells became to be observed, suggesting that initially infecting HIV-1 was degraded and/or replaced with replication-competent proviruses. After 3 days post-infection, EGFP+ cells were clearly visible and the proportion of EGFP−Gag+ cells turned to be increased, suggesting that progeny virus infection occurred. Because the CD4 expression levels were identical between EGFP−Gag+ cells and EGFP−Gag− cells throughout the culture period, Gag p24 ICS must have detected cells that had bound or internalized HIV-1.
Figure 5.
Evaluation of Gag p24 ICS for HIV-1-internalized and -bound cells. (A) Representative flow cytometric analyses of the HIV-1NL-E infection (NL-E) and mock (Mock) groups from three donors in which unstimulated primary CD4+ T cells were infected with HIV-1NL-E or not, respectively, and cultured for 5 days (including the initial 1 day culture with TCR-stimulation). The expression patterns of EGFP and Gag p24 are indicated in the pseudo-color plot profiles for the Dead−/CD3+/CD8−-gated cell fractions (upper panels). The expression level of CD4 in EGFP−Gag+ and EGFP−Gag− cells is indicated in the histograms by the blue and black lines, respectively (lower panels). (B) Representative flow cytometric analyses of Gag p24 staining. CEM–CCR5 cells were infected with HIV-1NL-E (NL-E), HIV-1AD8-E (AD8-E), or not (Mock). Immediately after spinoculation at 4°C, the cells were washed and fixed/permeabilized (fix/perm) or not (intact) prior to Gag p24 staining.
CEM–CCR5 cells, which are almost as susceptible to X4 and R5 HIV-1 fusion (data not shown), were used to confirm that Gag p24 ICS did indeed detect HIV-1-bound cells. Also, because it has been suggested that spinoculation at 25°C may induce HIV-1 fusion to the targeted cells (Dai et al., 2009), we tested Gag p24 ICS using CEM–CCR5 cells immediately after spinoculation with X4-tropic HIV-1NL-E or R5-tropic HIV-1AD8-E at 4°C. When cells were not fixed/permeabilized, no Gag p24+ cells were detected by flow cytometry (Figure 5B, upper panels); however, when cells were fixed/permeabilized, a substantial proportion of Gag+ cells was detectable in both the HIV-1NL-E and HIV-1AD8-E infection groups (Figure 5B, lower panels). Taken together, these results indicate that cells that have bound or internalized HIV-1 can be detected using flow cytometry for Gag p24 ICS.
The intensity of the HIV-1 fluorescent reporter signal depends on TCR-mediated activation levels
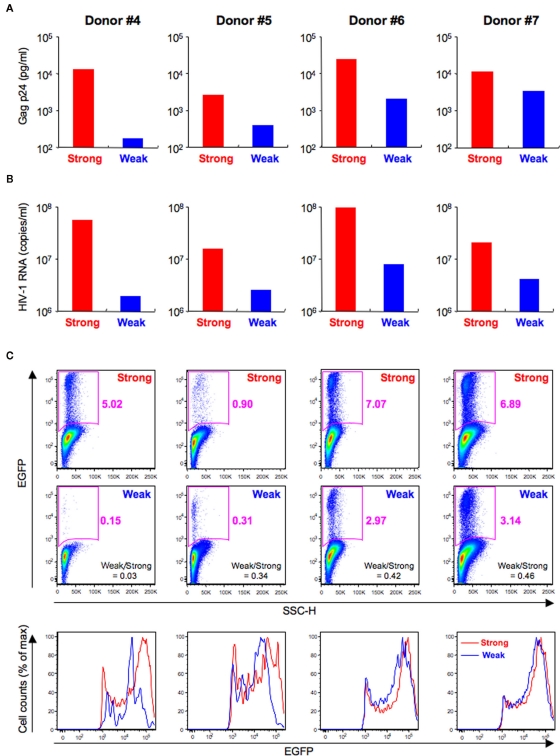
T cell receptors-mediated activation of HIV-1-infected CD4+ T cells increased productive viral replication, although the signaling pathway responsible may be different for X4 and R5 HIV-1 (Popik and Pitha, 2000). We investigated whether the intensity of the HIV-1 fluorescent reporter signal was affected by TCR-mediated activation levels. In this experiment, primary CD4+ T cells from four donors were individually pre-stimulated via the TCR for 4 (weak stimulation) or 24 h (strong stimulation), infected with HIV-1NL-E, and then cultured for a further 3 days. First, we confirmed that this experimental protocol allowed the preferential production of HIV-1NL-E upon strong stimulation in all donors by examining the cell culture supernatants by ELISA (Figure 6A) and real-time RT-PCR (Figure 6B). Flow cytometric analysis of intact cells showed that HIV-1NL-E+ (EGFP+) cells were more prevalent after strong stimulation than after weak stimulation, although the proportion of HIV-1NL-E+ cells varied among individuals (Figure 6C, upper and middle panels). The PMT voltage was optimized for EGFP to prevent excessive EGFP signaling (Figures 2 and 3). Of note, EGFP expression by HIV-1NL-E+ cells was lower in the weak stimulation group than in the strong stimulation group (as observed in donors #4 and #5), and EGFP expression in the weak stimulation group approached that in the strong stimulation group in parallel with the increase in the number of HIV-1NL-E+ cells (as observed in donors #6 and #7; Figure 6C, lower panels). Taken together, these results show that the intensity of the fluorescent reporter is highly correlated with the viral replication level.
Figure 6.
Differences of HIV-1 replication according to TCR-mediated activation levels. Primary CD4+ T cells from four donors (Donor #4–7) were pre-stimulated via TCR for 4 (Weak; blue) or 24 h (Strong; red) and then infected with HIV-1NL-E and cultured for 3 days. (A) ELISA for HIV-1 Gag p24 in the culture supernatants. (B) Quantitative real-time RT-PCR for HIV-1 RNA in the culture supernatants. (C) Flow cytometric analysis of intact cells showing pseudo-color plot profiles (Dead−/CD3+/CD8−-gated cell fraction; upper and middle panels) and histogram profiles (Dead−/CD3+/CD8−/EGFP+-gated cell fraction; lower panels).
Discussion
Flow cytometric analysis is a reliable and convenient method for detecting HIV-1-infected cells at a single cell level. Here, we studied the potential usefulness of several HIV-1 fluorescent reporters that have been published previously (Yamamoto et al., 2009). We examined whether they would be helpful for evaluating viral replication levels based on their fluorescence intensity. In this study, we used recombinant HIV-1 encoding either EGFP or DsRed to show that the fluorescence intensity of the EGFP and DsRed reporters was associated with the level of CD4 downmodulation (Figure 2). Furthermore, we showed that EGFP intensity was associated with the expression level of Gag p24 (Figure 4). These findings clearly indicate that fluorescent reporter intensity is useful for evaluating viral replication levels. To confirm this argument, we further compared the fluorescent reporter intensity of HIV-1-infected cells that were strongly or weakly stimulated via the TCR. As expected, higher levels of HIV-1 replication/production occurred in strongly stimulated cells from all the donors tested (Figure 6A,B). Although the proportion and EGFP intensity of the HIV-1-infected cells varied among individuals, this might be due to differing susceptibility to HIV-1 and/or TCR-stimulation. Thus, the variability in EGFP expression is rather favorable to our argument, as increased EGFP intensity was associated with an increase in the number of HIV-1-infected cells after weak stimulation (Figure 6C).
Although Gag p24 ICS is usually used for flow cytometric analysis of other markers, we showed that it can also be used to detect cells that have internalized or bound HIV-1 (Figure 5A,B). However, Gag p24 ICS did not appear sensitive enough to detect HIV-1-infected cells because some HIV-1-infected cells in which CD4 was moderately downmodulated were identified as positive for EGFP but negative for Gag p24 (Figure 4). Bosque and Planelles (2009) also identified a small population of such reporter-positive but Gag p24-negative cells by flow cytometry when CD4+ T cells were infected with EGFP-encoded DHIV incorporating a small out-of-frame deletion in the gp120-encoding area and pseudotyped with X4-tropic HIV-1LAI, and assumed that these cells were at an early stage of the infection process and did not display late viral proteins. Therefore, our own findings indicate that it is the HIV-1 fluorescent reporter, rather than Gag p24 staining, that reliably detects HIV-1-infected cells at different stages of infection in flow cytometry experiments.
It is known that maturation of DsRed for coloration is usually slower compared with EGFP (Bevis and Glick, 2002; Maruyama et al., 2004). When we focused on the HIV-1 dull fraction in Figure 2, we found that CD4 downmodulation was stronger in DsRed+ cells than in EGFP+ cells. These results suggest that the EGFP reporter is preferable to the DsRed reporter for detection of earlier stage of infection. Furthermore, the detrimental effect of fixation/permeabilization on fluorescent reporter intensity, particularly when using the DsRed reporter, should be noted (Figure 3). Although the detailed mechanism remains obscure, this may result from the lower fluorescence intensity of DsRed compared with EGFP. A similar phenomenon was described regarding fixation with 3% paraformaldehyde, which significantly decreases the fluorescence intensity of DsRed, although specific data were not provided (Weber et al., 2006). Regardless of the weakened signal, the EGFP reporter is still compatible with fixation/permeabilization because the proportion of EGFP+ cells was comparable between intact cells and fixated/permeabilized cells (Figure 3). Therefore, the EGFP reporter still maintains an advantage for analyses of cytokine/chemokine production and proliferation assays based on Ki-67 expression, for which ICS is necessary.
The HIV-1 fluorescent reporter has a potential application in molecular biology. In general, ICS-treated cells are not suitable for analysis using molecular biology techniques, since formaldehyde-based fixation (required for ICS) makes RNA extraction and reverse transcription and quantification problematic (Farragher et al., 2008) because of chemical cross-linking of proteins and nucleic acids (Kuykendall and Bogdanffy, 1992; Finke et al., 1993; Park et al., 1996), degradation of RNA (Bresters et al., 1994), and covalent modification of RNA via the addition of monomethylol groups to the bases (Masuda et al., 1999); therefore, by using the HIV-1 fluorescent reporter, HIV-1-infected cells can be sorted/purified without the need for fixation, allowing further characterization at a molecular level.
Given the usefulness of the HIV-1 fluorescent reporter shown here, it would also be very useful for investigating the mechanisms involved in the selective replication of R5 HIV-1 over X4 HIV-1 during the acute phase in vivo (Wolinsky et al., 1992; Zhu et al., 1993; van’t Wout et al., 1994) and in cell culture systems in vitro (Schweighardt et al., 2004; Roy et al., 2005). We previously developed an in vitro dual infection model using EGFP-encoded X4 HIV-1 (HIV-1NL-E) and DsRed-encoded R5 HIV-1 (HIV-1AD8-D) and showed that the increase in the proportion of X4 HIV-1-infected cells is dependent upon their activation level (Yamamoto et al., 2009). Furthermore, the results of the present study show that the fluorescence intensity of the reporter molecule can be used to assess the level of viral replication in infected cells; therefore, by focusing on the HIV-1-infected cells and the fluorescent reporter intensity in the dual infection model, the detailed mechanism(s) of HIV-1 infection/pathogenesis can be clarified. In this regard, we have been investigating the dynamics of HIV-1 infection in vivo using humanized mice infected simultaneously with EGFP-encoded X4 HIV-1 (HIV-1NL-E) and DsRed-encoded R5 HIV-1 (HIV-1AD8-D; Ishige et al., in preparation). We believe that the advantages of the recombinant HIV-1 fluorescent reporter will contribute to the further understanding of HIV-1 infection/pathogenesis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Kaori Okano for technical support. This work was supported in part by Grants from the Ministry of Education, Science, Sports, and Culture of Japan (Kazutaka Terahara), and the Ministry of Health, Labour, and Welfare of Japan (Kazutaka Terahara and Yasuko Tsunetsugu-Yokota).
References
- Berger E. A., Murphy P. M., Farber J. M. (1999). Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu. Rev. Immunol. 17, 657–700 10.1146/annurev.immunol.17.1.657 [DOI] [PubMed] [Google Scholar]
- Bevis B. J., Glick B. S. (2002). Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed). Nat. Biotechnol. 20, 83–87 10.1038/nbt0102-83 [DOI] [PubMed] [Google Scholar]
- Bosque A., Planelles V. (2009). Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113, 58–65 10.1182/blood-2008-07-168393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresters D., Schipper M. E., Reesink H. W., Boeser-Nunnink B. D., Cuypers H. T. (1994). The duration of fixation influences the yield of HCV cDNA-PCR products from formalin-fixed, paraffin-embedded liver tissue. J. Virol. Methods 48, 267–272 10.1016/0166-0934(94)90125-2 [DOI] [PubMed] [Google Scholar]
- Chalfie M. (1995). Green fluorescent protein. Photochem. Photobiol. 62, 651–656 10.1111/j.1751-1097.1995.tb08712.x [DOI] [PubMed] [Google Scholar]
- Cubitt A. B., Heim R., Adams S. R., Boyd A. E., Gross L. A., Tsien R. Y. (1995). Understanding, improving and using green fluorescent proteins. Trends Biochem. Sci. 20, 448–455 10.1016/S0968-0004(00)89099-4 [DOI] [PubMed] [Google Scholar]
- Dai J., Agosto L. M., Baytop C., Yu J. J., Pace M. J., Liszewski M. K., O’doherty U. (2009). Human immunodeficiency virus integrates directly into naive resting CD4+ T cells but enters naive cells less efficiently than memory cells. J. Virol. 83, 4528–4537 10.1128/JVI.01910-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky D. I., Wells M., Harrington R. D. (1996). Detection of HIV-1 infection with a green fluorescent protein reporter system. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 13, 308–313 10.1097/00042560-199612010-00002 [DOI] [PubMed] [Google Scholar]
- Farragher S. M., Tanney A., Kennedy R. D., Paul Harkin D. (2008). RNA expression analysis from formalin fixed paraffin embedded tissues. Histochem. Cell Biol. 130, 435–445 10.1007/s00418-008-0479-7 [DOI] [PubMed] [Google Scholar]
- Finke J., Fritzen R., Ternes P., Lange W., Dolken G. (1993). An improved strategy and a useful housekeeping gene for RNA analysis from formalin-fixed, paraffin-embedded tissues by PCR. Biotechniques 14, 448–453 [PubMed] [Google Scholar]
- Gervaix A., West D., Leoni L. M., Richman D. D., Wong-Staal F., Corbeil J. (1997). A new reporter cell line to monitor HIV infection and drug susceptibility in vitro. Proc. Natl. Acad. Sci. U.S.A. 94, 4653–4658 10.1073/pnas.94.9.4653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim R., Cubitt A. B., Tsien R. Y. (1995). Improved green fluorescence. Nature 373, 663–664 10.1038/373663a0 [DOI] [PubMed] [Google Scholar]
- Kar-Roy A., Dong W., Michael N., Li Y. (2000). Green fluorescence protein as a transcriptional reporter for the long terminal repeats of the human immunodeficiency virus type 1. J. Virol. Methods 84, 127–138 10.1016/S0166-0934(99)00122-6 [DOI] [PubMed] [Google Scholar]
- Kuykendall J. R., Bogdanffy M. S. (1992). Efficiency of DNA-histone crosslinking induced by saturated and unsaturated aldehydes in vitro. Mutat. Res. 283, 131–136 10.1016/0165-7992(92)90145-8 [DOI] [PubMed] [Google Scholar]
- Malim M. H., Emerman M. (2008). HIV-1 accessory proteins – ensuring viral survival in a hostile environment. Cell Host Microbe 3, 388–398 10.1016/j.chom.2008.04.008 [DOI] [PubMed] [Google Scholar]
- Maruyama M., Nishio T., Yoshida T., Ishida C., Ishida K., Watanabe Y., Nishikawa M., Takakura Y. (2004). Simultaneous detection of DsRed2-tagged and EGFP-tagged human beta-interferons in the same single cells. J. Cell. Biochem. 93, 497–502 10.1002/jcb.20203 [DOI] [PubMed] [Google Scholar]
- Masuda N., Ohnishi T., Kawamoto S., Monden M., Okubo K. (1999). Analysis of chemical modification of RNA from formalin-fixed samples and optimization of molecular biology applications for such samples. Nucleic Acids Res. 27, 4436–4443 10.1093/nar/27.22.4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcclure M. O., Sattentau Q. J., Beverley P. C., Hearn J. P., Fitzgerald A. K., Zuckerman A. J., Weiss R. A. (1987). HIV infection of primate lymphocytes and conservation of the CD4 receptor. Nature 330, 487–489 10.1038/330487a0 [DOI] [PubMed] [Google Scholar]
- O’doherty U., Swiggard W. J., Malim M. H. (2000). Human immunodeficiency virus type 1 spinoculation enhances infection through virus binding. J. Virol. 74, 10074–10080 10.1128/JVI.74.21.10074-10080.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page K. A., Liegler T., Feinberg M. B. (1997). Use of a green fluorescent protein as a marker for human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retroviruses 13, 1077–1081 10.1089/aid.1997.13.1077 [DOI] [PubMed] [Google Scholar]
- Park Y. N., Abe K., Li H., Hsuih T., Thung S. N., Zhang D. Y. (1996). Detection of hepatitis C virus RNA using ligation-dependent polymerase chain reaction in formalin-fixed, paraffin-embedded liver tissues. Am. J. Pathol. 149, 1485–1491 [PMC free article] [PubMed] [Google Scholar]
- Popik W., Pitha P. M. (2000). Inhibition of CD3/CD28-mediated activation of the MEK/ERK signaling pathway represses replication of X4 but not R5 human immunodeficiency virus type 1 in peripheral blood CD4(+) T lymphocytes. J. Virol. 74, 2558–2566 10.1128/JVI.74.6.2558-2566.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A. M., Schweighardt B., Eckstein L. A., Goldsmith M. A., Mccune J. M. (2005). Enhanced replication of R5 HIV-1 over X4 HIV-1 in CD4(+)CCR5(+)CXCR4(+) T cells. J. Acquir. Immune Defic. Syndr. 40, 267–275 10.1097/01.qai.0000176593.14172.52 [DOI] [PubMed] [Google Scholar]
- Saito A., Nomaguchi M., Iijima S., Kuroishi A., Yoshida T., Lee Y. J., Hayakawa T., Kono K., Nakayama E. E., Shioda T., Yasutomi Y., Adachi A., Matano T., Akari H. (2010). Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes Infect. 13, 58–64 10.1016/j.micinf.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Schweighardt B., Roy A. M., Meiklejohn D. A., Grace E. J., 2nd, Moretto W. J., Heymann J. J., Nixon D. F. (2004). R5 human immunodeficiency virus type 1 (HIV-1) replicates more efficiently in primary CD4+ T-cell cultures than X4 HIV-1. J. Virol. 78, 9164–9173 10.1128/JVI.78.17.9164-9173.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunetsugu-Yokota Y. (2008). “Transmission of HIV from dendritic cells to CD4+ T cells: a promising target for vaccination and therapeutic intervention,” in AIDS Vaccines, HIV Receptors and AIDS Research, ed. Kendow L. B. (New York: Nova Science Publishers, Inc.), 117–128 [Google Scholar]
- Tsunetsugu-Yokota Y., Akagawa K., Kimoto H., Suzuki K., Iwasaki M., Yasuda S., Hausser G., Hultgren C., Meyerhans A., Takemori T. (1995). Monocyte-derived cultured dendritic cells are susceptible to human immunodeficiency virus infection and transmit virus to resting T cells in the process of nominal antigen presentation. J. Virol. 69, 4544–4547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van’t Wout A. B., Kootstra N. A., Mulder-Kampinga G. A., Albrecht-Van Lent N., Scherpbier H. J., Veenstra J., Boer K., Coutinho R. A., Miedema F., Schuitemaker H. (1994). Macrophage-tropic variants initiate human immunodeficiency virus type 1 infection after sexual, parenteral, and vertical transmission. J. Clin. Invest. 94, 2060–2067 10.1172/JCI117560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J., Weberova J., Carobene M., Mirza M., Martinez-Picado J., Kazanjian P., Quinones-Mateu M. E. (2006). Use of a novel assay based on intact recombinant viruses expressing green (EGFP) or red (DsRed2) fluorescent proteins to examine the contribution of pol and env genes to overall HIV-1 replicative fitness. J. Virol. Methods 136, 102–117 10.1016/j.jviromet.2006.04.004 [DOI] [PubMed] [Google Scholar]
- Wolinsky S. M., Wike C. M., Korber B. T., Hutto C., Parks W. P., Rosenblum L. L., Kunstman K. J., Furtado M. R., Munoz J. L. (1992). Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255, 1134–1137 10.1126/science.1546316 [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Tsunetsugu-Yokota Y., Mitsuki Y. Y., Mizukoshi F., Tsuchiya T., Terahara K., Inagaki Y., Yamamoto N., Kobayashi K., Inoue J. (2009). Selective transmission of R5 HIV-1 over X4 HIV-1 at the dendritic cell-T cell infectious synapse is determined by the T cell activation state. PLoS Pathog. 5, e1000279. 10.1371/journal.ppat.1000279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T., Mo H., Wang N., Nam D. S., Cao Y., Koup R. A., Ho D. D. (1993). Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 261, 1179–1181 10.1126/science.7687073 [DOI] [PubMed] [Google Scholar]