Background: TDOR are ubiquitous and catalyze important cell redox reactions.
Results: Dtrx presents atypical physicochemical properties and a positive surface around its active site, suggesting a specificity for it(s) substrate(s).
Conclusion: Active site histidine plays an important role in the molecular mechanism of Dtrx catalysis.
Significance: Structural and functional studies of such atypical systems will give new insights into the TDOR catalytic mechanism.
Keywords: Disulfide, NMR, Protein Structure, Thiol, Thioredoxin, Desulfovibrio, pKa
Abstract
Cytoplasmic desulfothioredoxin (Dtrx) from the anaerobe Desulfovibrio vulgaris Hildenborough has been identified as a new member of the thiol disulfide oxidoreductase family. The active site of Dtrx contains a particular consensus sequence, CPHC, never seen in the cytoplasmic thioredoxins and generally found in periplasmic oxidases. Unlike canonical thioredoxins (Trx), Dtrx does not present any disulfide reductase activity, but it presents instead an unusual disulfide isomerase activity. We have used NMR spectroscopy to gain insights into the structure and the catalytic mechanism of this unusual Dtrx. The redox potential of Dtrx (−181 mV) is significantly less reducing than that of canonical Trx. A pH dependence study allowed the determination of the pKa of all protonable residues, including the cysteine and histidine residues. Thus, the pKa values for the thiol group of Cys31 and Cys34 are 4.8 and 11.3, respectively. The His33 pKa value, experimentally determined for the first time, differs notably as a function of the redox states, 7.2 for the reduced state and 4.6 for the oxidized state. These data suggest an important role for His33 in the molecular mechanism of Dtrx catalysis that is confirmed by the properties of mutant DtrxH33G protein. The NMR structure of Dtrx shows a different charge repartition compared with canonical Trx. The results presented are likely indicative of the involvement of this protein in the catalysis of substrates specific of the anaerobe cytoplasm of DvH. The study of Dtrx is an important step toward revealing the molecular details of the thiol-disulfide oxidoreductase catalytic mechanism.
Introduction
Thiol/disulfide oxidoreductases (TDOR)2 are ubiquitous in prokaryotes and eukaryotes and catalyze important redox reactions in the cell (1, 2). All of the members of this family share the thioredoxin fold consisting of a central β-sheet surrounded by four α-helices and an active site with two conserved cysteine residues that specify the biological activity of the protein (3). Despite these similarities, thiol/disulfide oxidoreductases can be subdivided according to their cellular location. Although the cytoplasmic members of this family such as thioredoxin (Trx) and glutaredoxin catalyze the reduction of disulfide bonds, the members of oxidizing cellular compartments, such as protein-disulfide isomerase (PDI) from the endoplasmic reticulum and disulfide bond proteins DsbA and DsbC from the bacterial periplasm, are catalysts of disulfide bond formation during folding of secreted proteins.
The CXXC active site of these proteins is essential for TDOR activity. The sequence of the XX dipeptide located between the cysteines in the active site motif is very important in controlling the redox properties of the protein (5–7). Within each subgroup the active site contains a conserved consensus sequence (Trx, CGPC; glutaredoxin, CPYC; PDI, CGHC; DsbA, CPHC; DsbC, CGYC).
In all thioredoxin-like proteins, the reactivity is influenced by the pKa value of the first active site cysteine residue and by the redox potential (E0′) of the disulfide bond. There is a remarkable correlation between the standard redox potential of these enzymes and their physiological role; indeed the members with the lowest redox potentials catalyze reducing processes in vivo (Trx, −270 mV (4); and glutaredoxin, −233 to −198 mV (5)), whereas the protein folding catalysts are strong oxidizing agents (PDI, −175 to −147 mV (6, 7); and DsbA, −163 to −80 mV (8)). Exceptions are the thioredoxin-like proteins anchored to the inner bacterial membrane. As an example, TlpA (thioredoxin-like protein A) from Bradyrhizobium japonicum exhibits a low redox potential (−259 mV) despite its periplasmic orientation and is required for cytochrome aa3 maturation (9). However, no high redox potential has ever been reported for cytoplasmic TDOR.
Thioredoxins are a group of small (12 kDa) proteins. They have been characterized from various prokaryotic and eukaryotic organisms (10). The thioredoxin system comprises a thioredoxin with the active site consensus sequence WCGPC (Trx), a thioredoxin reductase, and NADPH. The molecular catalytic mechanism actually proposed for the reduction of the oxidized substrate proteins by the canonical Trx involves a bimolecular nucleophilic substitution reaction. The reaction starts with a nucleophilic attack of the N-terminal thiol of the WCXXC motif on the disulfide bond of the target protein, releasing a free thiol and forming a mixed disulfide between Trx and its substrate. In the second step, the C-terminal thiol must be activated as a thiolate to allow the dissociation of the complex. For deprotonation of the C-terminal thiol, one hypothesis involves a conserved aspartate residue (11). Recently, it was suggested that when the N-terminal thiolate of Trx attacks its substrate disulfide to form a mixed disulfide complex, the leaving thiol group deprotonates the thiol of the C-terminal active site of Trx (12). Finally another hypothesis proposes that the cysteine is activated for its nucleophilic attack by hydrogen bonds between this residue and the backbone amide of the active site tryptophan (13).
In Desulfovibrio vulgaris Hildenborough, a Gram-negative sulfate-reducing bacteria, two cytoplasmic thioredoxin systems have been identified (14). The Trx1/TR1 system contains the ubiquitous thioredoxin with the active site consensus sequence WCGPC. The second system contains an atypical thioredoxin with a CPHC sequence at the active site identical to the DsbA motif and an unconventional thioredoxin reductase that uses preferentially NADH (14). The presence of these atypical proteins being restricted to Desulfovibrio organisms, they have been named desulfothioredoxin (Dtrx) and desulfothioredoxin reductase (15).
In this work, we have determined the disulfide isomerase and reductase activities and redox properties for the cytoplasmic CPHC active site. We have solved the structure of Dtrx and investigated the catalytic mechanism of this atypical enzyme. We identified important structural differences between Dtrx and the canonical bacterial Trx1 in the areas surrounding the catalytic sites.
EXPERIMENTAL PROCEDURES
Protein Production
The encoding sequence (DVU378) of desulfothioredoxin from D. vulgaris Hildenborough was cloned into expression vector pJF119-EH for production of a His-tagged protein at its C terminus.
pJF119-EH Dtrx plasmid with the desulfothioredoxin wild-type gene was used as DNA template in two separate PCRs to introduce the H33G mutation with two pair of primers (an internal mutagenic forward primer (H1: 5′-CCTGTGCCCGGGCTGCAAGAAC-3′) and an external reverse primer (C2: 5′-GCTTCTGCGTTCTGATTTAATCTG-3′); and an internal mutagenic reverse primer (H2: 5′-GTTCTTGCAGCCCGGGCACAGG-3′) and an external forward primer (C1: 5′-GCAGAAACGTGGCTGGCCTGG-3′)). The two PCR-purified fragments were mixed, denatured, and extended. The product was then amplified in a third PCR using C1 and C2 primers. The resulting fragments were digested with BamHI and EcoRI and cloned into the pJF119-EH vector.
Transformed Escherichia coli TG1 cells were grown in a M9 minimal medium containing 15NH4Cl (1 g/liter) and [13C]glucose (2 g/liter) as the sole nitrogen and carbon sources. Protein purification was achieved using nickel-nitrilotriacetic acid affinity chromatography and imidazole gradient (20–500 mm) in 500 mm NaCl and 100 mm phosphate buffer at pH 8. The purity of the protein sample was checked by SDS-PAGE.
NMR Spectroscopy
NMR spectra were recorded on Bruker Avance III 600 MHz spectrometer equipped with a TCI cryoprobe or a Bruker Avance III 500 MHz spectrometer equipped with a TXI probe. All of the experiments were carried out at 298 K. The spectra were processed using Topspin (Bruker).
Activity Assay
The ability of Trx1 and Dtrx, wild-type and mutant, to catalyze insulin reduction in the presence of DTT was determined as previously described (16). The reaction mixtures were prepared in cuvettes containing 130 μm insulin, 5 μm protein catalyst in different 0.1 m buffers (pH 4.22, 6.3, 7.06, 7.52, 8.11, and 8.98), and 2.5 mm EDTA. For Dtrx, we also used a 20 μm concentration. The reactions were started with the addition of 1 mm DTT. The rate of precipitation was monitored by recording the increased turbidity of the reaction mixture, which was measured at 650 nm every 30 s at 33 °C and using an Uvikon spectrophotometer. The noncatalyzed reduction of insulin by DTT was monitored in a control reaction.
An in vitro assay involving refolding of scrambled RNase A was used to monitor the oxidase activity of Trx1, Dtrx, DtrxH33G mutant, and DsbC (ATGen, Seongnam-SI, South Korea) (17). Disulfide-scrambled RNase A was produced by incubating 30 mg of native RNase A (Sigma) overnight at room temperature in 50 mm Tris-HCl, pH 8.5, in the presence of 6 m guanidinium chloride and 130 mm DTT. After acidifying the solution (addition of 1 μl of 11 m acetic acid), the DTT was removed by passing the sample through a desalting column. The RNase A concentration was then determined at 280 nm. The fully reduced sample was diluted to 0.5 mg·ml−1 in water at pH 6.7. Reshuffling of scrambled RNase A (15 μm) was carried out by incubation 15 min in 5 mm potassium phosphate buffer, pH 7.0, with 60 μm protein sample. The RNase activity was assayed by analysis of RNA hydrolysis after 30 min of reaction by NMR. For the determination of a percentage of the RNase A activity, the mean intensity of several isolated peaks in one-dimensional NMR spectrum of RNA was used relative to the RNA spectrum in the presence of native RNase A. The RNA spectrum in the presence of ScRNase A is used as blank.
An in vivo assay involving refolding of PalB (lipase B from Pseudozyma antarctica) was used to monitor the isomerase activity of Trx1 and Dtrx. DNA fragments containing the leaderless palB gene were PCR-amplified using PrimeSTAR HS DNA polymerase (TAKARA), the primer pair (P1/P3 or P2/P3), and P. antarctica chromosomal DNA as template. One PCR product was cloned into the restriction sites SacI/XbaI of the pJF119EH vector. The second PCR product was used to obtain by PCR a palB fusion construct with Trx1 and Dtrx ORFs and cloned into pJF119EH vector with EcoRI/XbaI (P1, 5′-CCGAGCTCATGCTACCTTCCGGTTCGGACCCTG-3′; P2, 5′-CGGTAGTGGTTCTGGGCTACCTTCCGGTTCGGACC-3′; and P3, 5′-GCTCTAGATCAGGGGGTGACGATGCCGGAGC-3′).
Rosetta-gami 2 and TG1 E. coli cells were transformed with pJF119EH constructs. The cells were grown in LB medium containing 100 μg/ml ampicillin at 310 K. When the cell density reached ∼0.8 A600, PalB, DTrx-PalB and Trx1-PalB expressions were induced by the addition of 1 mm isopropyl β-d-thiogalactopyranoside at 37 °C overnight. PalB activity was qualitatively evaluated by the area and the transparency of the halo formed on the tributyrin agar plate (LB agar plate containing 1% emulsified tributyrin). The PalB-producing strains were transferred onto the plate and incubated at room temperature until the halos developed.
Determination of Redox Potential
For NMR spectroscopy, two samples of 0.3 mm Dtrx and 0.6 mm DtrxH33G were dialyzed against 50 mm potassium phosphate buffer, pH 7.0, supplemented with 4 mm GSSG. Dtrx and DtrxH33G were titrated with GSH in the ranges 0–30 and 0–90 mm, respectively. 15N-1H HSQC spectra were recorded upon titration with reduced or oxidized glutathione. For the determination of the redox potential, the intensities of several isolated peaks in each NMR spectrum were obtained from Topspin and plotted versus the half-cell potential of glutathione. Redox potential was obtained by fitting the experimental curve against a sigmoidal decay (logistic) function. Subsequently, E0′ values obtained for individual peaks were averaged.
The half-cell potential of glutathione for the reaction GSSG + 2H+ + 2e− → 2GSH was calculated according to the Nernst equation,
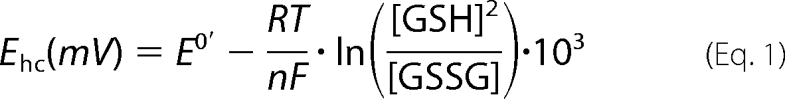 |
where R is the universal gas constant (8.3145 J K−1 mol−1), T is the temperature in Kelvin, n = 2 for the two-electron reduction, and F is the Faraday constant (9.6485·104 C mol−1).
The standard (midpoint) potential (E0′) was defined as the potential at 50% of the maximal resonance intensity. For the GSH/GSSG pair, E0′ was previously determined to be −240 mV at 298 K, pH 7.0 (5).
pKa Determination
NMR experiments were carried out on samples containing 1 mm concentration protein with and without DTT. The behavior of the 13C chemical shifts in the protein as a function of pH was monitored using a two-dimensional CBCACO experiment (18). Assignment of these chemical shifts was verified for each pH using a three-dimensional HNCO experiment. Chemical shift values as a function of pH were analyzed according to a single titration curve as shown in Equation 2 (19),
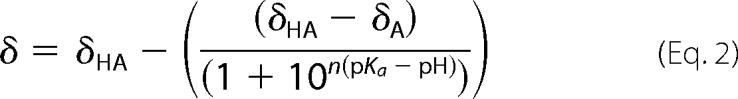 |
where δ is the observed chemical shift at a given pH, δHA and δA are the chemical shifts for the various protonated forms of the protein, and n is the number of protons transferred.
Structure Calculation
The NMR sample contained 1 mm protein concentration (90% H2O, 10% D2O) in 100 mm NaCl, 50 mm phosphate buffer, pH 5.5. For experiments regarding the reduced form of the protein, the intramolecular disulfide bond of Dtrx was reduced by adding DTT to a final concentration of 10 mm, under argon atmosphere. The spectra were analyzed with CARA (20) on the basis of the previously published backbone amide and side chain resonances assignment (15). The approximate interproton distances were obtained from the two-dimensional NOESY, 13C NOESY-HSQC, and 15N NOESY-HSQC spectra. For structure calculations, 1846 restraints were used for the reduced Dtrx and 2108 restraints for the oxidized form. The mixing time was 150 ms for all of the NOESY experiments. A series of 15N-1H HSQC spectra was acquired on a sample freshly dissolved in 2H2O to identify the slowly exchanging amides. Amides that had not exchanged after 1 h were located in regions of defined secondary structures, based on the NOE data, and were restrained to form HN-CO hydrogen bonds, using the distance restraints of 2.7–3.0 Å for O-N and 1.8–2.0 Å for O-HN, respectively. For structure calculations, 48 restraints were used for backbone hydrogen bonds for the reduced Dtrx and 70 restraints for the oxidized form. 156 backbone ϕ and ψ dihedral supplemental restraints were derived from TALOS using as input the 1Hα, 13Cα, 13Cβ, 13C′, and 15N chemical shifts (21).
Input data and structure calculation statistics are summarized in supplemental Table S1. The accuracy of the NMR models could be assessed based on the criteria for successful structure calculation using the program CYANA (25). The 20 lowest energy (total energy) structures chosen for the final structural ensemble were subjected each to restrained molecular dynamics using the Amber 4.1 force field within the SANDER module of Amber 10. The water molecules were stripped off, and energy terms were calculated for the protein using AMBER. The nonbonded interaction cutoff was 15 Å for the restrained MD runs. The structure coordinates have been deposited in the Protein Data Bank under accession numbers 2L6D and 2L6C for the reduced and oxidized Dtrx, respectively.
RESULTS
Dtrx Activities
The particular CPHC sequence at the Dtrx active site opens the question of the enzymatic function of this protein. Therefore, we have examined the redox activity of this atypical protein using in vitro assays.
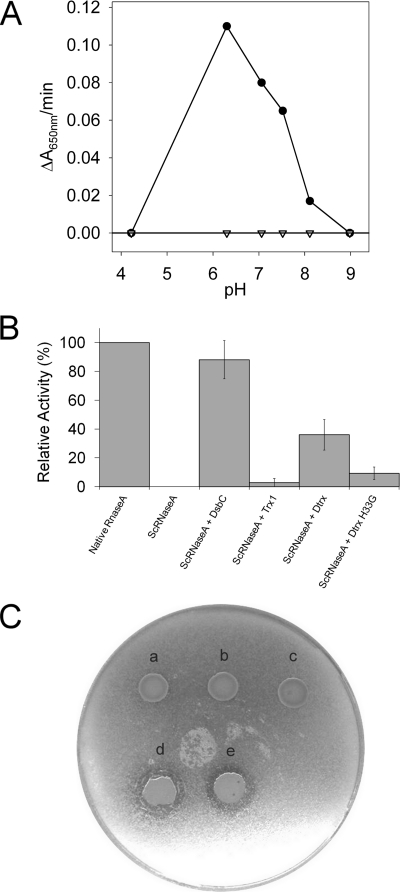
First, we used the insulin reduction assay at different pH levels. In this assay, oxidoreductase enzymes reduced the interchain disulfide bond of insulin, causing precipitation of the insoluble insulin β-chain. Precipitation was monitored as an increase in absorbance at 650 nm. We have realized this assay with both Trx1 and Dtrx at pH ranging from 4 to 9. As expected, we found that Trx1 is as active as E. coli Trx. Surprisingly, Dtrx is unable to reduce insulin even at concentrations 10 times greater (Fig. 1A). It is the first time that a thioredoxin-related protein does not show activity in insulin assay. However, Dtrx demonstrates a reductase activity on artificial substrate as dithiobis(nitrobenzoic acid) (14).
FIGURE 1.
Dtrx activities. A, insulin reduction assay by Trx1 and Dtrx as a function of pH. The disulfide reductase activity was determined by using the insulin reduction assay. The assay was performed at 306 K with 2 μm Dtrx (triangle) or 2 μm Trx1 (black circle). The catalyzed reduction of insulin (100 μm) was followed by measuring an increase in absorbance at 650 nm and evaluated at different pH levels: 4.22, 6.3, 7.06, 7.52, 8.11, and 8.98. The absorbance caused by the nonenzymatic insulin reduction by DTT (1 mm) is substracted. B, scrambled RNase A (ScRNase) refolding assay: yield of RNase A activity of native RNase A and reshuffling of ScRNase A after incubation with DsbC, Trx1, Dtrx, and Dtrx H33G mutant. For the determination of a percentage of the RNase A activity, the mean intensity of several isolated peaks in RNA spectrum was used relative to the RNA spectrum in the presence of native RNase A. The RNA spectrum in presence of ScRNase A is used as blank. The error analysis of the data points was performed using Excel software. C, study of in vivo activity. Qualitative visualization of PalB activity using tributyrin plate for recombinant E. coli TG1 and Rosetta-gami as host cells. Different expression systems were observed: pJF119EH PalB (a), pJF119EH Dtrx-PalB (b), pJF119EH Trx1-PalB (c) in E. coli TG1 and pJF119EH PalB (d) and pJF119EH Dtrx-PalB (e) in E. coli Rosetta-gami.
We investigated the ability of Dtrx to catalyze disulfide isomerization. We measured the capacity of Dtrx to isomerize, or shuffle, incorrect disulfides of scrambled RNase A at pH 7 (Fig. 1B). After incubation of scrambled RNase A and Dtrx (molar ratio 1:4), we have followed the RNase activity by observing the digestion of RNA using 1H NMR spectra (supplemental Fig. S1). We have used the DsbC isomerase as a positive control. We found that, under our experimental conditions, after 15 min of incubation, the sample containing Dtrx and Trx1 yielded 40 and 5% active RNase A, respectively. DsbC yielded 85% active RNase A. The weak activity of Trx1 is similar to E. coli Trx1, and Dtrx disulfide isomerase activity is comparable with that of E. coli DsbA oxidase (22).
To verify the in vivo disulfide isomerase activity of Dtrx, we have explored the functional expression of PalB in the cytoplasm of E. coli using fusion tag techniques (23). Indeed, PalB has three intramolecular disulfide bonds potentially associated with its folding process and required for developing the bioactivity. The fusion tag technique was explored by constructing two PalB fusions, Trx1 and Dtrx tag. We have compared the expression performance using E. coli TG1 and Rosetta-gami strains having a reducing and oxidizing cytoplasm, respectively, as host cells. PalB activity was qualitatively evaluated by the area and transparency of the halo formed on the tributyrin agar plate (Fig. 1C). No visible halos developed for Dtrx-PalB fusion. Contrary to DsbA, Dtrx was not able to enhance PalB folding in the E. coli cytoplasm by disulfide bond formation (23).
Dtrx Redox Potential
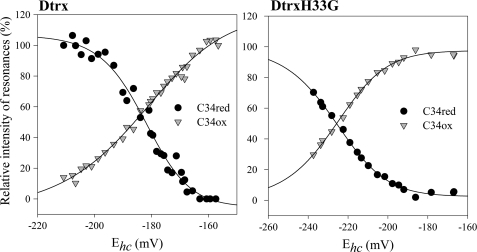
The next step of our study was to compare the redox potential of Dtrx and canonical Trx. The measurement of the redox potential of Dtrx was obtained by 15N-1H HSQC NMR experiments (15). We utilized GSH and GSSG as the redox couple, which has a standard potential of −240 mV at pH 7.0 and 298 K (5). The intensities of the NH resonances of the oxidized form of Dtrx decreased upon the addition of reduced glutathione. At the same time, the intensities of the NH resonances of the reduced form increased. The measurement of the NMR signal intensities resulted in a sigmoidal transition curve. Several chemical shift changes associated with the reduction of the catalytic cysteines of the protein were observed (supplemental Fig. S2). Sigmoidal curves were fitted successfully for the Cys34 resonances (Fig. 2). The redox potential for the cysteines of the active site at pH 7.0 was −181.3 ± 1.2 mV. In conclusion, this value indicated that Dtrx was more oxidizing than canonical Trx (−270 mV).
FIGURE 2.
Titration of the redox potential of the disulfide bond of Dtrx and DtrxH33G. Changes in NMR signal intensities for the NH resonances of Cys34 under oxidizing or reducing conditions were reported in the function of half-cell potential of glutathione. Gray triangles and black circles represent signal intensity in the oxidized and reduced states, respectively. Redox potentials were calculated using the Nernst equation from the ratio of concentrations of reduced and oxidized glutathione. Experimental data were fitted against a sigmoidal decay (logistic) function.
pKa of Dtrx Protonable Residues
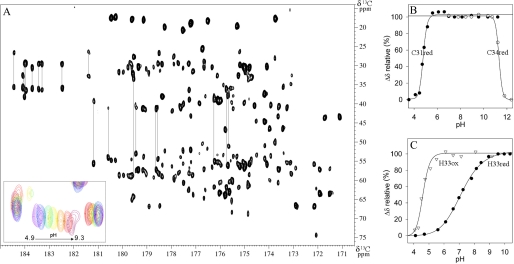
Because there is generally a correlation between the redox potential of the active site and the pKa of the cysteine residues, we have investigated the pKa of all the residues of Dtrx. The knowledge of the protonation state for the residues involved in the reaction mechanism is highly important (28, 29), but the pKa values are not always easy to determine experimentally for all residues (19). This is the first time that a proton-less NMR spectrum (CBCACO) (Fig. 3A) was used to follow the pH dependence of 13Cβ chemical shift of all residues. In the reduced form, the titration curves in Fig. 3B revealed pKa values of 4.8 and 11.3 for the thiol group of Cys31 and Cys34 residues, respectively. These values were different from those already reported for canonical Trx: 7.5 for the first cysteine and 9.5 for the second (11). The pKa of the active cysteine thiol group reflected the stabilization of the thiolate anion. To understand this peculiar property and get some insights into the Dtrx catalytic mechanism, we have determined the pKa value of all protonable residues in the protein (supplemental Fig. S3). Only the pKa values of His33 from the CPHC active site presented an atypical value and changed strongly between the two redox states of Dtrx, these values being 7.2 and 4.6 for the imidazole group in the reduced and oxidized states, respectively (Fig. 3C). It is the first time that pKa values of the histidine residues found in the CPHC active site motif have been experimentally determined. These data are of particular interest to get precise atomic views of the catalytic site in both redox states of the protein.
FIGURE 3.
pKa determination of all Dtrx ionizable residues. A, 600 MHz two-dimensional CBCACO spectrum of reduced Dtrx at 298 K, pH 5.7, showing the cross-peaks for the Cα-CO and Cβ-CO of all residues of the protein. Cross-peaks for the Cα-Cγ and Cβ-Cγ of Asn and Asp, and for the Cβ-Cδ and Cγ-Cδ of Gln and Glu are also visible and connected by lines. The inset shows a close-up view of the pH-dependent chemical shift variations for the Cβ-CO of Asp21 (pH 4.9 (pink), 5.6 (red), 6.1 (orange), 6.6 (yellow), 7 (green), 7.8 (blue), and 9.3 (purple)). B, pKa determination of the nucleophilic cysteine Cys31 (black circle) and cysteine Cys34 (white circle) in the reduced form of Dtrx. C, pKa determination of the histidine His33 in the oxidized form (white triangle) and reduced form (black circle) of Dtrx. The pH-dependent chemical shift variation of the Cβ carbons was measured, normalized, and fitted to one apparent pKa value using the Henderson-Hasselbach equation.
Role of His33 in Dtrx Catalytic Mechanism
To confirm a potential role of His33 in the particular properties of Dtrx, we have mutated this residue into a glycine residue. This mutant protein (DtrxH33G) was produced and purified as the wild type. The 1H,15N HSQC spectrum was recorded and compared with the Dtrx spectrum (supplemental Fig. S4). This comparison shows that the mutation does not modify the structure of the protein because only residues close to the mutation undergo weak chemical shift variations. Next, we have determined the activities and the redox potential of the DtrxH33G mutant. In the same experimental conditions as the wild type, we have studied the ability of DtrxH33G to reduce insulin and, like Dtrx, DtrxH33G is unable to reduce insulin. It is thus not the presence of a histidine residue in the active site that induces this particular property of the protein. However, the DtrxH33G mutant shows a loss of the capacity of the protein to isomerize, or shuffle, incorrect disulfides of scrambled RNase A yielding 10% active RNase A only (Fig. 1B and supplemental Fig. S1). The histidine residue or the physicochemical properties induced by this histidine are essential for Dtrx to catalyze disulfide isomerization. The determination of the redox potential of DtrxH33G shows effectively that His33 plays an important role in the Dtrx properties (Fig. 2). The redox potential of the active site cysteines at pH 7.0 for DtrxH33G (−226.4 ± 0.6 mV) is 45 mV more reducing than for the wild type. This value is intermediate between Dtrx (−181.3 mV) and canonical Trx (−270 mV) redox potentials.
Dtrx Structure Analysis
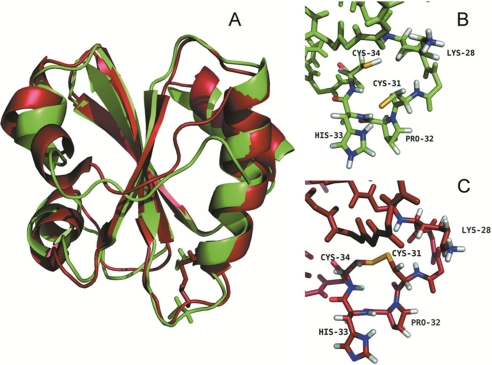
From our studies, the activities, the redox potential, and the pKa values of Dtrx are significantly different from a canonical Trx. To gain insight into these atypical properties of Dtrx and to propose a molecular catalytic mechanism, we have determined the three-dimensional structure of the enzyme in the two redox states. The reduced and oxidized structures of Dtrx were calculated at pH 5.5 using interproton nOe-derived distance restraints in combination with the dihedral angle (21) and hydrogen bond restraints (supplemental Table S1). We used the pKa values to define the protonation state of the histidine and cysteine residues in the structure refinement. The resulting ensembles of solutions consisting of the 20 lowest energy structures of the oxidized and the reduced Dtrx are shown in supplemental Fig. S5. These ensembles have a backbone root mean square deviation relative to the average structure of 0.59 ± 0.11 and 0.94 ± 0.15 Å over the polypeptide chain for the oxidized and the reduced Dtrx, respectively. Detailed structure statistics are shown in supplemental Table S1.
Oxidized and reduced Dtrx adopt a typical thioredoxin fold (3). The oxidized form consists of a four-stranded twisted central β-sheet (residues 52–58 (β1), 20–27 (β2), 74–81 (β3), and 83–90 (β4)) surrounded by four α-helices (residues 12–17 (α1), 33–46 (α2), 61–69 (α3), and 93–104 (α4)) (Fig. 4A). The overall structure of the reduced Dtrx is similar to the one of the oxidized form. Dtrx displays high three-dimensional similarities to thioredoxins from other species such as E. coli Trx1 and the thioredoxin fold domain of other thiol disulfide oxidoreductases such as E. coli DsbA and DsbC (30, 31) (supplemental Fig. S6).
FIGURE 4.
Three-dimensional structure of Dtrx in both redox states. A, overlay of the three-dimensional solution structures of the reduced (green) and oxidized (red) forms of Dtrx calculated with CYANA (21). The NMR sample contained 1 mm protein concentration (90% H2O, 10% D2O) in 100 mm NaCl, 50 mm phosphate buffer, pH 5.7, at 290 K. For reduced Dtrx, the intramolecular disulfide bond was reduced by adding DTT to a final concentration of 10 mm, under argon atmosphere. The Dtrx typical thioredoxin fold is represented in cartoon, and the side chains of the cysteine residues are shown in sticks. B, local conformations of the active site in the reduced form of Dtrx. C, local conformations of the active site in the oxidized form of Dtrx. The active site residues Cys31, Pro32, His33, and Cys34 are shown and labeled. Sulfur atoms are shown in yellow, hydrogen atoms are in white, and nitrogen and oxygen atoms are in blue and red, respectively. The figures were generated using the PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.
With the exception of the disulfide bond, the main structural differences between the two redox states of Dtrx concern the helix α1 because of the number of restraints used in the structure calculation. The active site (31CPHC34) of Dtrx is formed by a protruding loop between strand β2 and the N termini of helix α2. Between the two redox states, the cysteine side chain undergoes change in orientation (Fig. 4, B and C). In the dithiol form, the S atoms are at a mean distance of 4.8 Å. Moreover, a difference in orientation of the histidine His33 is observed, positioning the cationic imidazole facing the thiolate anion of the reactive cysteine Cys31 at a mean distance of 3.8 Å. In 14 of 20 conformers of the NMR structure, the thiolate anion of Cys31 forms two hydrogen bonds with the backbone amide and the HNδ of His33.
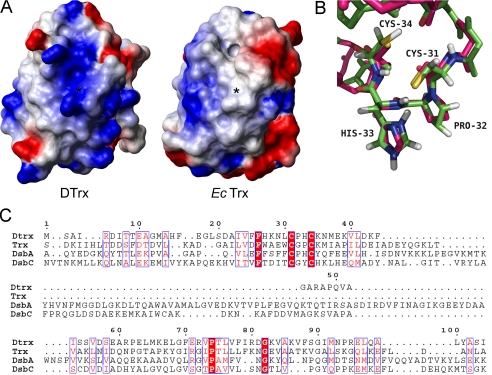
In the reduced Dtrx form, the Cys31 side chain is exposed at the protein surface, and the Cys34 side chain is pointing toward the interior of the protein, as is the case in the active site of reduced E. coli Trx1. The electrostatic surface of Dtrx shows a different charge repartition compared with canonical Trx. Indeed, we observe a positive surface instead of the highly conserved hydrophobic patch found around the canonical active site (Fig. 5, A and B). The active site of Dtrx is perfectly superimposable to the one of DsbA. Pro32 and His33 residues occupy the same position when compared with the same residues in DsbA (1A2J) (Fig. 5C).
FIGURE 5.
Comparative structural analysis of Dtrx. A, electrostatic surface potential representations of reduced Dtrx and reduced E. coli Trx, with blue representing basic residues, red representing acidic residues, and white representing neutral residues. The orientations of Dtrx and Trx are similar. The position of the sulfur atom of the N-terminal cysteine is indicated by a star. The hydrophobic patch observed in Trx is not conserved in Dtrx. Surface calculations were made using MolMol. B, superimposition of the redox active site of the reduced form of Dtrx (green) and E. coli DsbA (pink) in stick representation. The active site residues are labeled. C, sequence alignment of Dtrx, E. coli Trx, E. coli DsbA, and E. coli DsbC. Identical residues are in red boxes. Conserved residues are shown in red. The alignment was prepared with TCoffee (34) and ESPript (11).
Thioredoxin and DsbA proteins generally contain the conserved residues Asp26 and Glu24, respectively, which have been described as involved in the activation of the second cysteine (Fig. 5D). As for DsbC, these residues are not found in Dtrx, and no acidic residue is found in the near environment of Cys34. These structural data obtained on Dtrx active site and the striking electrostatic surface potential around the active site strongly suggest a specific function of Dtrx in the anaerobe cytoplasm.
DISCUSSION
Properties of Dtrx
Based on sequence homologies, desulfothioredoxin (Dtrx) from D. vulgaris Hildenborough has been identified as a new member of the thioredoxin superfamily. Dtrx contains a particular active site consensus sequence, CPHC, found in the periplasmic DsbA. Dtrx does not show any disulfide reductase activity, being unable to reduce insulin. To date, no protein of the thioredoxin superfamily has shown comparable properties. According to the study of the DtrxH33G mutant, this property is not related to the particular active site of this protein. One explanation is the particular structural properties of Dtrx. The first step of the redox mechanism of Trx is the noncovalent interaction of the oxidized substrate with the hydrophobic surface of the active Trx (10). DsbA contains a helical insertion that forms a hydrophobic patch on the molecular surface, as well as a hydrophobic groove near the active site (32, 33). Rinaldi et al. (24) have observed a negative electrostatic surface of Xylella fastidiosa DsbA2, indicating that this protein versus DsbA has different substrate specificities. It is to be noticed that Dtrx contains a positive surface and no hydrophobic patch around its active site. These differences are the main characteristic of Dtrx structure and may induce a strong specificity for its substrate(s).
An in vitro assay involving refolding of scrambled RNase A showed an unusual disulfide isomerase activity of Dtrx. Considering the nature and composition of the sequence in the active site, this activity can be explained. In fact, CXXC affects the standard redox potential of the particular proteins. In E. coli, the strongest reducing cytosolic Trx (CGPC) has a redox potential of ΔE0′ = −270 mV (25), and the strongest oxidizing agents, the periplasmic DsbA (CPHC) and DsbC (CGYC), have a redox potential of ΔE0′ = −122 mV (26). The redox potential value of human PDI (CGHC) is an intermediate, ΔE0′ = −175 mV (27). The redox potential observed for the cysteine active site of Dtrx at pH 7.0 is −181.3 mV. This characteristic shows that Dtrx is closer to oxidizing enzymes than canonical reductases and even closer to PDI isomerase. This property is in part due to the presence of histidine residue in the active site. Indeed, DtrxH33G lost the ability to catalyze the disulfide isomerization and presents a redox potential closer to disulfide reductases (−226.4 mV).
The pKa values of 4.8 and 11.3 obtained respectively for the thiol group of Cys31 and Cys34 residues, are different from those found in all canonical Trx. In E. coli Trx1, the pKa of the thiol group is 7.5 for the first cysteine and 9.5 for the second cysteine in the consensus site (11). The pKa of this thiol group reflects the stabilization of the thiolate anion of the accessible cysteine residue. Strong stabilization of the thiolate generates a low pKa value and low stability for the disulfide bond; this phenomenon was observed for Dtrx. The instability of the disulfide bond is characteristic of oxidase or isomerase proteins because it is necessary for their catalytic mechanism (28). In summary, these physicochemical properties of Dtrx are in agreement with the unusual disulfide isomerase activity found in in vitro assays for Dtrx.
However, in vivo, Dtrx did not present any isomerase activity in the E. coli cytoplasm, because it has been observed for DsbA. Indeed, Xu et al. (23) showed that a functional expression of PalB in the cytoplasm of E. coli appeared to be limited by disulfide bond formation, and a DsbA fusion tag was functional to enhance PalB folding in cytoplasm. Despite the observed in vitro disulfide isomerase activity, Dtrx was not able to perform this activity in vivo in E. coli cytoplasm. Is it a problem of cytoplasmic conditions or of substrate specificity?
The cytoplasm of most organisms is a highly reducing environment, in which protein cysteines are maintained in thiol/thiolate form. However, in extremophile organisms, disulfide bond formation in cytosolic proteins is supposed to increase their thermodynamic stability. Several protein-disulfide oxidoreductases in the disulfide-rich thermophiles were identified and could have dual functions in the cytoplasm, as oxidase and isomerase (29). Therefore we can suppose that oxidase activity could exist in the cytoplasm. Dtrx contains a positive surface and no hydrophobic patch around its active site. These differences are the main characteristic of Dtrx structure and may induce a strong specificity for its substrate(s) and explain the absence of oxidation of lipase in vivo, where the ratio Dtrx:lipase is 1:1 in the cell.
Is Dtrx Really a Thioredoxin?
Considering the absence of in vitro reductase activity of Dtrx and its oxidizing physicochemical properties (redox potential, pKa values of the active site), a potential thioredoxin function is questionable. Nevertheless, glutaredoxin, which is a good reducing agent, was also reported to have oxidizing redox properties provided by its low pKa of the active cysteine (30). Conversely, thioredoxins, which are potent reducing agents, can also act as oxidizing agents under particular conditions. Indeed, E. coli thioredoxin can be translocated to the periplasm and then partially replaces the activity of DsbA in promoting the formation of disulfide bonds (31). Thioredoxin can function as a reductase or oxidase or isomerase, mainly depending on the redox environment.
For Dtrx, several aspects support a reductase activity: (i) the three-dimensional structure of Dtrx revealed an identical fold to canonical thioredoxins without a supplementary domain; (ii) the gene DVU_0378 encoding Dtrx is included in a polycistronic unit counting nine other genes, including DVU_0377, which encodes a thioredoxin reductase (desulfothioredoxin reductase) and is able to reduce Dtrx but not any other thioredoxin (14); and (iii) Dtrx has a reductase activity on dithiobis(nitrobenzoic acid) substrate (14).
Catalytic Mechanism of Dtrx
It is the first time that the pKa values of the histidine of CPHC active site were determined. These atypical values vary from 7.2 to 4.6 for the reduced to the oxidized Dtrx, respectively.
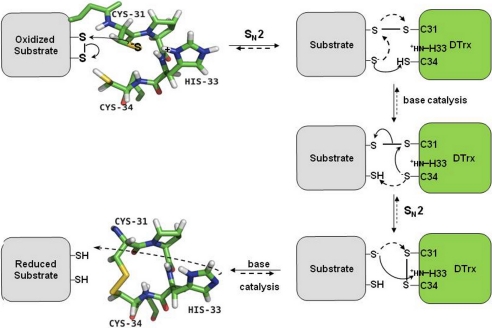
On the basis of these results, we propose a catalytic mechanism for the reduction of the oxidized substrate by the reduced Dtrx (Fig. 6). The nucleophilic cysteine in the reduced Dtrx is deprotonated by water molecules because of its low pKa (4.8). The Cys31 thiolate of Dtrx nucleophilically attacks a disulfide sulfur atom of the substrate, leading to the formation of the so-called mixed disulfide intermediate in which Dtrx and substrate are covalently bound via a new disulfide bond. Next, the Cys34 thiol group of Dtrx is deprotonated. However, the mechanism through which the buried Cys34 is deprotonated and activated to perform the intramolecular nucleophilic attack on Cys31 is still uncertain in canonical Trx. One hypothesis is that an aspartic acid conserved in all canonical Trx is responsible for this deprotonation (32). It is to be noticed that a double alanine mutation of the conserved Asp26 and Lys57 in Trx and Glu24 and Lys58 in DsbA did not change the pKa of cysteines in both enzymes. These data support the hypothesis that these residues are not involved in the properties of the dithiol active center (33). A second hypothesis involves the thiolate leaving group, which establishes S−…H-S hydrogen bonds with the thiol group of the buried cysteines (12). However, the mutation of the second cysteine of the substrate shows a dissociation of the intermolecular complex (13). Finally, in a last hypothesis, the cysteine is proposed to be activated for its nucleophilic attack by hydrogen bonds between this residue and the backbone amides of the tryptophan active site and of the N-terminal cysteine (13). These hydrogen bonds explain the stabilization of thiolate group but not the deprotonation of this group. In Dtrx, the aspartic acid is not conserved, and analysis of pKa value of all residues does not present any acid group available for the activation of Cys34. Therefore, we centered the mechanism around the second hypothesis where the thiolate of the substrate attacks the Cys34 sulfur atom or any acid group of the substrate. The protonated His33 would stabilize by a salt bridge successively Cys31 and Cys34 thiolates. In the second step, Cys34 thiolate would attack the Cys31 sulfur atom involved in the disulfide bond, causing the rupture of the latter. At the same time, the imidazole group in the oxidized Dtrx being deprotonated (pKa 4.6), we propose that the thiolate of the reduced substrate would attack His33 and induce the release of the products of the reaction, an oxidized enzyme with deprotonated His33 and a reduced dithiol substrate.
FIGURE 6.
Model of the Dtrx catalytic mechanism. Substrate reduction (solid lines) by Dtrx occurs in four steps. In the first step, the Cys31 thiolate of Dtrx nucleophilically attacks sulfur atom of the substrate disulfide. In the second step, the thiolate of the substrate produces a base attack on Cys34 sulfur atom. Next, the Cys34 thiolate attacks the Cys31 sulfur atom involved in the disulfide bond, causing the rupture of the latter. At the same time, the imidazole group of His33 is deprotonated by a base attack of the thiolate of the reduced substrate. This reaction produces an oxidized Dtrx and a reduced dithiol substrate. Substrate oxidation (dotted lines) by Dtrx occurs in four steps. First, the deprotonated His33 produces a base attack on the first cysteine of the substrate. After, this activated cysteine of the substrate nucleophilically attacks the sulfur atom of Cys31 of the oxidized Dtrx, leading to the formation of a disulfide-linked complex between Dtrx and the substrate. In the next step, the second cysteine of the substrate is deprotonated probably by the Cys34 thiolate of Dtrx and attacks the sulfur atom of the substrate cysteine, which is disulfide-bonded with Cys31 of Dtrx. This reaction results in the formation of a disulfide bond in the substrate and the reduction of Dtrx with a protonated His33.
As for all thioredoxins, this mechanism is reversible. DsbA contains the same CPHC sequence motif, but no data are available on the pKa values for the imidazole group of this protein. We suggest that the protonation state could be identical to Dtrx. In DsbA catalysis, the two-step bimolecular nucleophilic substitution mechanism described in the literature (34) can be completed by a first step substrate activation. In this case, the imidazole group of histidine being deprotonated (pKa 4.6) attacks the thiol group of the reduced substrate and induces the activation of the substrate with the formation of a thiolate cysteine. Effectively, in E. coli, it is unlikely that all of the 300 potential DsbA substrates present reactive cysteines (low pKa). They most likely have to be activated by the deprotonated active site histidine of DsbA. In DsbA, mutations of the histidine located at the active site, diminished drastically the oxidase activity affecting the destabilization of the oxidized form of the enzyme (28, 35). According to the authors, the mutation affects the destabilization of the oxidized form of the enzyme. We think that the histidine mutant will affect not only the stability of disulfide bond but especially the activation of the substrates.
In conclusion, the new thioredoxin Dtrx from anaerobe D. vulgaris Hildenborough presents several unusual structural features and contains a particular active site consensus sequence, CPHC, which gives oxidizing properties. Our data suggest a particular specificity for its substrates, and one cannot exclude that this specificity may be limited to one of the gene products of the operon it belongs to. Further studies of these gene products will be essential to determine the physiological function of Dtrx. The knowledge of the protonation state for the residues involved in the reaction mechanism is highly important, and the pKa values for all residues of the protein were determined for the first time. pKa value determination allowed us to define the important function of the histidine residue at the catalytic site. Genome analysis of many bacteria reveals unusual thioredoxins. The structural and functional studies of such atypical systems will give new insights into TDOR catalytic mechanism.
Supplementary Material
Acknowledgments
We thank Dr. A. Dolla and Dr. B. Burlat for helpful discussions and Dr. P. Barré for critical reading of the manuscript.

This article contains supplemental Table S1 and Figs. S1–S6.
The atomic coordinates and structure factors (codes 2L6D and 2L6C) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TDOR
- thiol-disulfide oxidoreductases
- Dsb
- disulfide bond protein
- Dtrx
- desulfothioredoxin
- PDI
- protein-disulfide isomerase
- Trx
- thioredoxin
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Holmgren A., Björnstedt M. (1995) Thioredoxin and thioredoxin reductase. Methods Enzymol. 252, 199–208 [DOI] [PubMed] [Google Scholar]
- 2. Rietsch A., Beckwith J. (1998) The genetics of disulfide bond metabolism. Annu. Rev. Genet 32, 163–184 [DOI] [PubMed] [Google Scholar]
- 3. Martin J. L. (1995) Thioredoxin: a fold for all reasons. Structure 3, 245–250 [DOI] [PubMed] [Google Scholar]
- 4. Mössner E., Huber-Wunderlich M., Glockshuber R. (1998) Characterization of Escherichia coli thioredoxin variants mimicking the active-sites of other thiol/disulfide oxidoreductases. Protein Sci. 7, 1233–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Aslund F., Berndt K. D., Holmgren A. (1997) Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein-protein redox equilibria. J. Biol. Chem. 272, 30780–30786 [DOI] [PubMed] [Google Scholar]
- 6. Chambers J. E., Tavender T. J., Oka O. B., Warwood S., Knight D., Bulleid N. J. (2010) The reduction potential of the active site disulfides of human protein disulfide isomerase limits oxidation of the enzyme by Ero1α. J. Biol. Chem. 285, 29200–29207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kersteen E. A., Raines R. T. (2003) Catalysis of protein folding by protein disulfide isomerase and small-molecule mimics. Antioxid. Redox Signal. 5, 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Quan S., Schneider I., Pan J., Von Hacht A., Bardwell J. C. (2007) The CXXC motif is more than a redox rheostat. J. Biol. Chem. 282, 28823–28833 [DOI] [PubMed] [Google Scholar]
- 9. Capitani G., Rossmann R., Sargent D. F., Grütter M. G., Richmond T. J., Hennecke H. (2001) Structure of the soluble domain of a membrane-anchored thioredoxin-like protein from Bradyrhizobium japonicum reveals unusual properties. J. Mol. Biol. 311, 1037–1048 [DOI] [PubMed] [Google Scholar]
- 10. Holmgren A. (1985) Thioredoxin. Annu. Rev. Biochem. 54, 237–271 [DOI] [PubMed] [Google Scholar]
- 11. Jeng M. F., Holmgren A., Dyson H. J. (1995) Proton sharing between cysteine thiols in Escherichia coli thioredoxin: implications for the mechanism of protein disulfide reduction. Biochemistry 34, 10101–10105 [DOI] [PubMed] [Google Scholar]
- 12. Carvalho A. T., Swart M., van Stralen J. N., Fernandes P. A., Ramos M. J., Bickelhaupt F. M. (2008) Mechanism of thioredoxin-catalyzed disulfide reduction: activation of the buried thiol and role of the variable active-site residues. J. Phys. Chem. B 112, 2511–2523 [DOI] [PubMed] [Google Scholar]
- 13. Roos G., Foloppe N., Van Laer K., Wyns L., Nilsson L., Geerlings P., Messens J. (2009) How thioredoxin dissociates its mixed disulfide. PLoS Comput. Biol. 5, e1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pieulle L., Stocker P., Vinay M., Nouailler M., Vita N., Brasseur G., Garcin E., Sebban-Kreuzer C., Dolla A. (2011) Study of the thiol/disulfide redox systems of the anaerobe Desulfovibrio vulgaris points out pyruvate:ferredoxin oxidoreductase as a new target for thioredoxin 1. J. Biol. Chem. 286, 7812–7821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Garcin E. B., Bornet O., Pieulle L., Guerlesquin F., Sebban-Kreuzer C. (2010) 1H, 13C and 15N backbone and side-chain chemical shift assignments for oxidized and reduced desulfothioredoxin. Biomol. NMR Assign. 4, 135–137 [DOI] [PubMed] [Google Scholar]
- 16. Holmgren A. (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 17. Hillson D. A., Lambert N., Freedman R. B. (1984) Formation and isomerization of disulfide bonds in proteins: protein disulfide-isomerase. Methods Enzymol. 107, 281–294 [DOI] [PubMed] [Google Scholar]
- 18. Bertini I., Felli I. C., Gonnelli L., Pierattelli R., Spyranti Z., Spyroulias G. A. (2006) Mapping protein-protein interaction by 13C-detected heteronuclear NMR spectroscopy. J. Biomol. NMR 36, 111–122 [DOI] [PubMed] [Google Scholar]
- 19. Jeng M. F., Dyson H. J. (1996) Direct measurement of the aspartic acid 26 pKa for reduced Escherichia coli thioredoxin by 13C NMR. Biochemistry 35, 1–6 [DOI] [PubMed] [Google Scholar]
- 20. Keller R. L. J. (2004) Computer aided resonance assignment tutorial. Cantina [Google Scholar]
- 21. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 22. Shouldice S. R., Cho S. H., Boyd D., Heras B., Eser M., Beckwith J., Riggs P., Martin J. L., Berkmen M. (2010) In vivo oxidative protein folding can be facilitated by oxidation-reduction cycling. Mol. Microbiol. 75, 13–28 [DOI] [PubMed] [Google Scholar]
- 23. Xu Y., Yasin A., Tang R., Scharer J. M., Moo-Young M., Chou C. P. (2008) Heterologous expression of lipase in Escherichia coli is limited by folding and disulfide bond formation. Appl. Microbiol. Biotechnol. 81, 79–87 [DOI] [PubMed] [Google Scholar]
- 24. Rinaldi F. C., Meza A. N., Guimarães B. G. (2009) Structural and biochemical characterization of Xylella fastidiosa DsbA family members: new insights into the enzyme-substrate interaction. Biochemistry 48, 3508–3518 [DOI] [PubMed] [Google Scholar]
- 25. Krause G., Lundström J., Barea J. L., Pueyo de la Cuesta C., Holmgren A. (1991) Mimicking the active site of protein disulfide-isomerase by substitution of proline 34 in Escherichia coli thioredoxin. J. Biol. Chem. 266, 9494–9500 [PubMed] [Google Scholar]
- 26. Wunderlich M., Glockshuber R. (1993) Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 2, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lundström J., Holmgren A. (1993) Determination of the reduction-oxidation potential of the thioredoxin-like domains of protein disulfide-isomerase from the equilibrium with glutathione and thioredoxin. Biochemistry 32, 6649–6655 [DOI] [PubMed] [Google Scholar]
- 28. Grauschopf U., Winther J. R., Korber P., Zander T., Dallinger P., Bardwell J. C. (1995) Why is DsbA such an oxidizing disulfide catalyst? Cell 83, 947–955 [DOI] [PubMed] [Google Scholar]
- 29. Ladenstein R., Ren B. (2006) Protein disulfides and protein disulfide oxidoreductases in hyperthermophiles. FEBS J. 273, 4170–4185 [DOI] [PubMed] [Google Scholar]
- 30. Fernandes A. P., Holmgren A. (2004) Glutaredoxins: glutathione-dependent redox enzymes with functions far beyond a simple thioredoxin backup system. Antioxid. Redox. Signal. 6, 63–74 [DOI] [PubMed] [Google Scholar]
- 31. Huber D., Cha M. I., Debarbieux L., Planson A. G., Cruz N., López G., Tasayco M. L., Chaffotte A., Beckwith J. (2005) A selection for mutants that interfere with folding of Escherichia coli thioredoxin-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 102, 18872–18877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chivers P. T., Raines R. T. (1997) General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry 36, 15810–15816 [DOI] [PubMed] [Google Scholar]
- 33. Carvalho A. T., Fernandes P. A., Ramos M. J. (2006) Determination of the DeltapKa between the active site cysteines of thioredoxin and DsbA. J. Comput. Chem. 27, 966–975 [DOI] [PubMed] [Google Scholar]
- 34. Kadokura H., Beckwith J. (2010) Mechanisms of oxidative protein folding in the bacterial cell envelope. Antioxid. Redox Signal. 13, 1231–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guddat L. W., Bardwell J. C., Glockshuber R., Huber-Wunderlich M., Zander T., Martin J. L. (1997) Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci. 6, 1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.