Background: Glycogenin autoglucosylation, required to prime glycogen glucopolymerization, can be produced by the monomeric and dimeric forms of the enzyme.
Results: Glycogenin intramonomer glucosylation produced full autoglucopolymerization, and intrasubunit glucosylation was necessary to complete dimer autoglucosylation.
Conclusion: Glycogenin dimerization is not required for full autoglucosylation.
Significance: De novo glycogen biosynthesis can be sustained by monomeric glycogenin.
Keywords: Enzyme Mechanisms, Glycogen, Glycosylation, Glycosyltransferases, Protein Structure, Autoglucosylation, Glycogenin
Abstract
Initiation of glucose polymerization by glycogenin autoglucosylation at Tyr-194 is required to prime de novo biosynthesis of glycogen. It has been proposed that the synthesis of the primer proceeds by intersubunit glucosylation of dimeric glycogenin, even though it has not been demonstrated that this mechanism is responsible for the described polymerization extent of 12 glucoses produced by the dimer. We reported previously the intramonomer glucosylation capability of glycogenin without determining the extent of autoglucopolymerization. Here, we show that the maximum specific autoglucosylation extent (MSAE) produced by the non-glucosylated glycogenin monomer is 13.3 ± 1.9 glucose units, similar to the 12.5 ± 1.4 glucose units measured for the dimer. The mechanism and capacity of the dimeric enzyme to carry out full glucopolymerization were also evaluated by construction of heterodimers able to glucosylate exclusively by intrasubunit or intersubunit reaction mechanisms. The MSAE of non-glucosylated glycogenin produced by dimer intrasubunit glucosylation was 16% of that produced by the monomer. However, partially glucosylated glycogenin was able to almost complete its autoglucosylation by the dimer intrasubunit mechanism. The MSAE produced by heterodimer intersubunit glucosylation was 60% of that produced by the wild-type dimer. We conclude that both intrasubunit and intersubunit reaction mechanisms are necessary for the dimeric enzyme to acquire maximum autoglucosylation. The full glucopolymerization capacity of monomeric glycogenin indicates that the enzyme is able to synthesize the glycogen primer without the need for prior dimerization.
Introduction
Previous reports on α-1,4-glucan-protein formation, proposed for the initiation of glycogen biosynthesis (1), and on the existence of retina glycogen as a protein-bound polysaccharide (2) led to the discovery of the autoglucosylating enzyme glycogenin (3, 4). Glycogenin autoglucosylation results in the formation of a glycogenin-bound Tyr-194-linked α-1,4-oligoglucan (5, 6), required to prime de novo glycogen biosynthesis (7, 8). The synthesized branched polysaccharide remains linked to the Tyr residue of glycogenin, forming a proteoglycogen molecule (9–11). Dimeric glycogenin, as it exists in the crystallized protein (12), is considered to be the autoglucosylating enzyme form (13). The glycogenin-linked oligoglucan product (analyzed by MS) was previously shown to have an average polymerization degree of 12 (6). The reaction kinetics exhibited by a heterodimer formed by two glycogenin mutants, one lacking glucosylating activity and the other without the acceptor Tyr-194 residue, led an intersubunit reaction mechanism being proposed for autoglucosylation (14). We demonstrated previously that monomeric glycogenin, as it is present in solution at submicromolar concentrations, is able to autoglucosylate by an intramolecular mechanism (15). However, the extent of glucopolymerization produced by the monomer was not determined.
Given the intramonomer glucosylation capacity, an intrasubunit glucosylation mechanism might also be involved in dimer autoglucosylation. Furthermore, enzyme dimerization might not be necessary to produce a fully glucopolymerized primer. The requirement of glycogenin to initiate de novo polysaccharide biosynthesis for the storage of metabolic energy reserve emphasizes the important role of the enzyme and the need to understand fully the reaction mechanism by which the primer is synthesized with the maximum degree of glucopolymerization. The requirement that glycogenin must reach a certain, as yet undetermined degree of glucosylation to prime further glucosylation by glycogen synthase has been reported previously (16). Thus, maximum polymerization of the synthesized glycogenin-bound oligoglucan might be necessary for the acceptor requirement of glycogen synthase or the branching enzyme to be able to carry out further glucosylation or the proposed formation of the first branch before elongation (5), respectively.
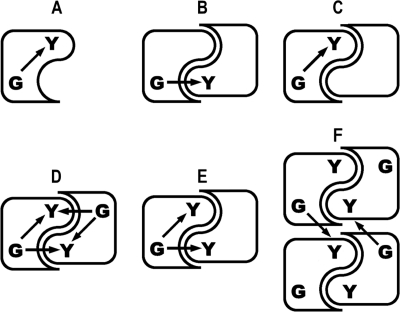
In this study, we analyzed the capacity of the glycogenin monomer to produce fully glucopolymerized Tyr-bound primer (Scheme 1A) and of heterodimers to autoglucosylate exclusively by an intersubunit or intrasubunit mechanism (Scheme 1, B and C, respectively). We also considered the possibility of both intrasubunit and intersubunit reaction mechanisms acting in combination in dimer autoglucosylation (Scheme 1D) and of tetrameric glycogenin interdimer glucosylation (Scheme 1F), as proposed previously (6, 13).
SCHEME 1.
Arrows indicate the glucose transfer from UDP-glucose bound in the active site (G) to Tyr-194 (Y) in the diagrammed glycogenin monomer (A), heterodimers with only intersubunit (B) or intrasubunit (C) glucosylation capacity, dimers with both intrasubunit and intersubunit glucosylation capacity (D and E), and the tetramer with interdimer glucosylation capacity (F) as proposed previously (6, 13).
EXPERIMENTAL PROCEDURES
Materials
Escherichia coli strain Rosetta(DE3) and vector pET15b were purchased from Novagen. Escherichia coli strain CGSC 4997, a galactose-minus strain lacking UDP-glucose pyrophosphorylase activity, was obtained from the E. coli Genetic Stock Center of the Department of Molecular, Cellular, and Developmental Biology at Yale University. The rabbit muscle 38-kDa glycogenin clone used as the PCR template was a gift from Dr. P. J. Roach (Indiana University School of Medicine). UDP-[14C]glucose (320 mCi/mmol) was purchased from the Instituto de Investigaciones Bioquímicas Fundación Leloir (Buenos Aires, Argentina). HisTrap FF and Superdex 75 columns were obtained from GE Healthcare. Except where indicated, all other reagents were from Sigma.
Wild-type and Mutant Recombinant Glycogenins
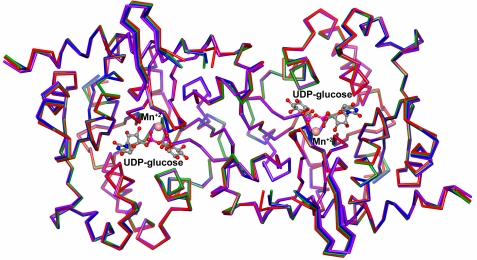
The active rabbit muscle glycogenins used in this work were the partially glucosylated full-length 38-kDa species (WT-38) and the non-glucosylated (apo-Δ270) and partially glucosylated (WT-Δ270) species truncated at residue 270. Truncation of glycogenin at residue 270 resulted in a more stable 31-kDa protein, without loss of activity (13, 14). The following autoglucosylation-inactive mutants were prepared: Y194F-Δ270, a double D159S/Y194F-Δ270 mutant lacking both glucosylating activity and acceptor capacity, and the T82M-Δ270 mutant recently described as causing a human deficiency in priming glycogen synthesis (8). We determined that the recombinant T82M-Δ270 mutant was devoid of autoglucosylating, transglucosylating, and UDP-glucose-hydrolyzing activities, showing a three-dimensional structure similar to that of apo-Δ270.4 It has been demonstrated previously that substitution of Tyr-194 with Phe and of Asp-159 with Ser eliminates the glycogenin acceptor capacity and glucosylating activity, respectively (14), without affecting the three-dimensional structures (13). The structures of Y194F-Δ270 and D159S/Y194F-Δ270 were solved, showing a high overall structural similarity to wild-type apo-Δ270, with root mean square deviations of 0.22 and 0.31 Å on the main chain atoms, respectively. The superimposition of these structures with the described three-dimensional structures of T82M-Δ2704 highlighted the similarity between them, despite their loss of activity (Fig. 1).
FIGURE 1.
Alignment of Y194F-Δ270 (green), T82M-Δ270 (red), and D159S/Y194F-Δ270 (purple) glycogenin mutants with wild-type apo-Δ270 enzyme (blue). The structures were superimposed with the LSQKAB program (23), and the figure was prepared using the CCP4MG program (24).
Mutagenesis, Expression, and Purification
Glycogenin truncated at residue 270 was obtained as described previously (15). Briefly, the cDNA corresponding to the fragment of interest was inserted in-frame in the pET15b vector downstream of the His6 tag-encoding sequence of the plasmid. The resulting plasmid was used as a template to generate all of the mutants by site-directed mutagenesis utilizing the QuikChange kit (Stratagene). For preparation of the glucosylated WT-Δ270 species and mutants, the appropriate plasmid was transformed into E. coli strain Rosetta(DE3). The truncated non-glucosylated species (apo-Δ270) was prepared by expression in E. coli strain CGSC 4997, previously lysogenized for site-specific integration of a T7 RNA polymerase gene using a λDE3 lysogenization kit (Novagen). Typically, 1 liter of culture was grown at 37 °C to absorbance values of 0.8–1.0 before being induced with 0.15 mm isopropyl β-d-thiogalactopyranoside overnight at 20 °C. The cells were collected by centrifugation, resuspended in 20 mm Tris-HCl buffer (pH 7.5) containing 0.5 m NaCl and 10 mm imidazole (column buffer), and homogenized using an EmulsiFlex-C3 device (Avestin, Inc., Ottawa, Canada). For purification of the His6-glycogenins, the clarified lysate was passed through a HisTrap FF column (1 ml). The column was then washed to give base-line UV absorbance with column buffer, and the proteins were eluted with an imidazole gradient (0–250 mm). Fractions containing glycogenin were dialyzed against 20 mm Tris-HCl buffer (pH 7.5) containing 0.24 m sucrose and stored at −20 °C.
Crystallization, Data Collection, and Structure Determination
Y194F-Δ270 and D159S/Y194F-Δ270 glycogenin mutants were concentrated to 10 mg/ml and crystallized in 12% (w/v) PEG monomethyl ether 5000, 0.1 m MES (pH 6.5), and 0.2 m ammonium sulfate using the hanging drop vapor diffusion method at 4 °C as described previously (13). Crystals were cryoprotected by soaking in a solution containing 70% mother liquor and 30% glycerol before being flash-frozen. Data were collected at protein crystallography beamline D03B-MX1 (wavelength, 1.430 Å) of the Laboratório Nacional de Luz Síncrotron (LNLS; Campinas, Brazil) at 100 K using a Marresearch CCD detector. Y194F-Δ270 and D159S/Y194F-Δ270 mutant crystals belonged to space group I222, with one subunit in the asymmetric unit. The data were processed and scaled using MOSFLM (17) and Scala (18). Subsequent data analysis was performed using the CCP4 suite of programs for crystallographic computing (19). The diffraction data statistics of the selected data sets are summarized in supplemental Table S1. As crystals of both glycogenin mutants are isomorphous with the wild-type ones, the refined structure of apo-Δ2704 without solvent molecules was used as a starting model for the refinement using the program REFMAC5 (20). After the Ser-159 and Phe-194 side chains had been introduced, the models were subjected to successive rounds of refinement alternated with manual inspection and model building with the programs REFMAC5 and Coot (21). Solvent molecules were added to the models in the final stages of refinement based on examination of the difference density maps.
Gel Filtration Chromatography
Gel filtration was carried out on a Superdex 75 column (24 ml) as described previously (15). The column was operated at 0.5 ml/min, 0.2-ml fractions were collected, and the radioactivity was counted.
Autoglucosylation
The glycogenin monomer, dimer, and heterodimers were subjected to 14C autoglucosylation by incubation at 30 °C with 320 μm UDP-[14C]glucose (20 mCi/mmol) in the presence of 0.1 m MES (pH 7.0), 5 mm MnSO4, and 1 mg/ml BSA in total volumes of 10 μl for the dimer and 100 μl for the glycogenin heterodimer and monomer. Unless indicated otherwise, the incubation time was 60 min. 14C autoglucosylation was measured by precipitation, washing the labeled protein with trichloroacetic acid, and radioactivity counting as described (22). Unless indicated otherwise, the results represent the mean values of three independent experiments, and error bars show S.D.
Gel Electrophoresis
The glycogenin monomer and heterodimers were 14C-autoglucosylated as described above, and aliquots were subjected to SDS-PAGE on 10% acrylamide resolving gels with 3% stacking gels. After running, the gels were stained with Coomassie Blue and subjected to autoradiography.
Mass Spectrometry
Electrospray ionization-MS was carried out on a Micromass Q-Tof mass spectrometer (Waters, Milford, MA) at the W. M. Keck Biotechnology Resource Laboratory at Yale University. Fully autoglucosylated glycogenin was prepared for MS analysis by incubation of 4.0 μm apo-Δ270 (320 pmol) with 320 μm UDP-glucose for 60 min as described above. Both non-autoglucosylated and autoglucosylated apo-Δ270 (320 pmol) were precipitated and then washed twice with cold acetone. The dried protein residue was reconstituted with 10 μl of 60% acetonitrile containing 0.1% formic acid before been injected into the mass spectrometer.
RESULTS
Maximum Autoglucopolymerization Extent of Glycogenin Monomer
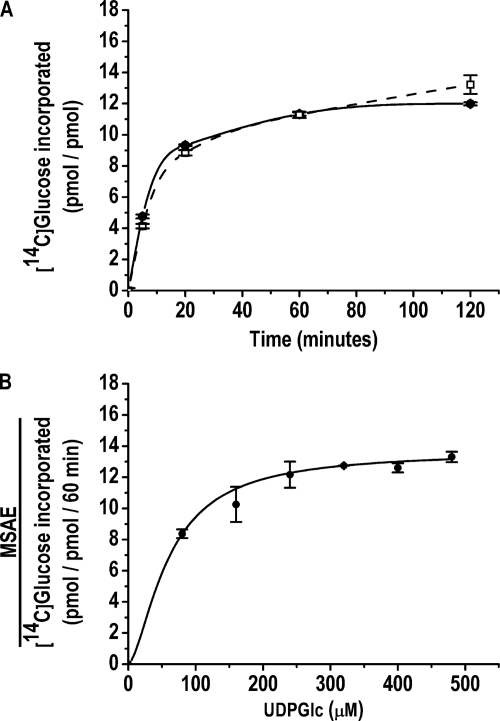
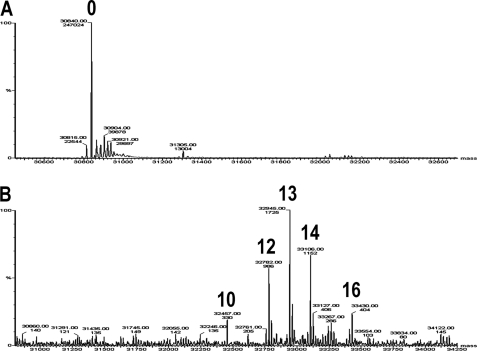
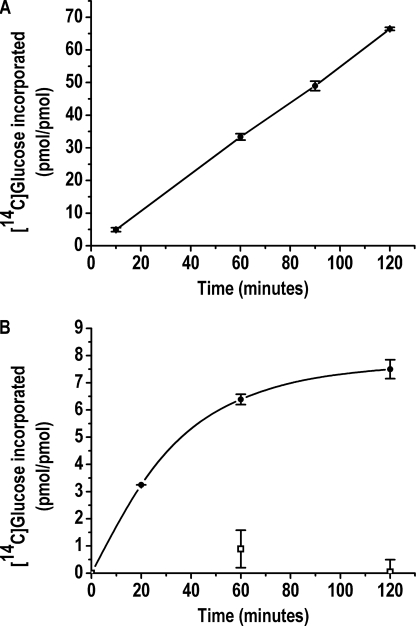
Glycogenin was assayed at two concentrations, 0.04 and 4.0 μm, which corresponded to the monomeric and dimeric enzyme forms based on the apparent dimer dissociation constant of 0.85 μm calculated previously (15). The full glucopolymerization capacities of 0.04 μm monomeric and 4.0 μm dimeric apo-Δ270 were determined by measuring the maximum specific autoglucosylation extent (MSAE)5 produced by incubation with UDP-[14C]glucose for 60 min, the time at which specific autoglucosylation reached a plateau (Fig. 2A). It was determined previously that 320 μm UDP-[14C]glucose is the concentration that assures saturation with the donor substrate for autoglucosylation and the accompanying sugar nucleotide-hydrolyzing activity (15) at the highest enzyme concentration used (Fig. 2B). The MSAE was 13.3 ± 1.9 glucose units for the monomer and 12.5 ± 1.4 glucose units for the dimer. The extent of glucopolymerization of dimeric glycogenin was corroborated by MS analysis. The deconvoluted spectrum showed glycogenin-bound glucans of 10, 12, 13, 14, and 16 glucose units, which, according to the relative abundance of each glucosylated species, resulted in an average polymerization degree of 13 (Fig. 3), similar to that of the average chain length described previously (6). The low concentration at which the enzyme exists as a monomer was an obstacle for determining the autoglucopolymerization degree by electrospray ionization-MS. Nevertheless, the agreement between the maximum specific 14C autoglucosylation extent measured for the dimer and the average extent determined by MS analysis supported the validity of the MSAE determined for the monomeric glycogenin form.
FIGURE 2.
Time course of monomer- and dimer-specific autoglucosylation (A) and dimer MSAE against UDP-glucose concentration (B). The 14C autoglucosylation of 0.04 μm monomeric (□) and 4.0 μm dimeric (●) apo-Δ270 glycogenins was carried out as described under “Experimental Procedures.”
FIGURE 3.
Deconvoluted mass spectra of fully autoglucosylated dimeric apo-Δ270 glycogenin. The apo-Δ270 species was analyzed by MS as described under “Experimental Procedures” before (A) and after (B) autoglucosylation by incubation of 4.0 μm glycogenin with unlabeled UDP-glucose for 60 min as described in the legend to Fig. 2. The different glucosylated glycogenin species are labeled 10, 12, 13, 14, and 16 according to the number of Tyr-linked glucose units. The relative abundance of each species was 7.2, 21.4, 37.5, 25.0, and 8.8%, respectively.
Extent of Glucopolymerization Produced by Dimer Intersubunit and Intrasubunit Glucosylation
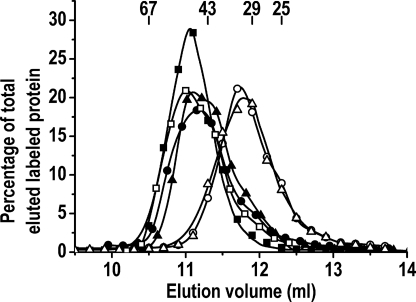
As mentioned above, mixing two autoglucosylation-inactive mutants, one lacking the glucosylation capacity and the other the acceptor capacity, resulted in the formation of a heterodimer that showed autoglucosylation kinetics consistent with an intersubunit glucosylation mechanism (14). However, the maximum extent of glucopolymerization produced by dimer intersubunit glucosylation was not determined. Our work on dimer autoglucosylation was focused on measuring the MSAE of heterodimers able to glucosylate exclusively by an intersubunit or intrasubunit reaction. The former was accomplished by measuring the autoglucosylation extent of the heterodimer formed between T82M-Δ270, a mutant recently described as causing a human deficiency in priming glycogen synthesis (8), and Y194F-Δ270 (Scheme 1B). The latter was undertaken by measuring the MSAE of the heterodimer formed by the D159S/Y194F-Δ270 double mutant, lacking glucosylating activity and acceptor capacity, and the autoglucosylation-active Δ270 species (Scheme 1C). Heterodimer formation was carried out by mixing and equilibration of a monomer species (0.04 μm) with the dimeric form of another species (3.96 μm). This concentration difference of 2 orders of magnitude favored the shift in the equilibrium toward the conversion of the monomer species into a heterodimer. The formation of the heterodimer was verified by gel filtration chromatography (Fig. 4). A complete shift toward a lower elution volume of labeled glycogenin was observed when 0.1 μm monomeric apo-Δ270, labeled by partial 14C autoglucosylation, was mixed with 20 μm dimeric D159S/Y194F-Δ270 double mutant and passed through a Superdex 75 column after equilibration. A similar elution shift was observed when the D159S/Y194F-Δ270 mutant was replaced in the mixture with the Y194F-Δ270 or T82M-Δ270 mutant (Fig. 4). This was consistent with the conversion of the labeled monomeric glycogenin to the heterodimer with any of the three mutants. The specificity of the heterodimer formation was corroborated by mixing similar proportions of 14C-glucosylated glycogenin and albumin, which resulted in no elution shift of the labeled enzyme.
FIGURE 4.
Gel filtration of glycogenin monomer, homodimer, and heterodimers. Monomeric apo-Δ270 (0.1 μm) was labeled in a 5-min incubation with 10 μm UDP-[14C]glucose and passed through a Superdex 75 column before (○) and after mixing and equilibration for 20 min with 20 μm T82M-Δ270 (■), Y194F-Δ270 (▴), D159S/Y194F-Δ270 (●), or albumin (△). 14C-Glucosylated dimeric apo-Δ270 (20 μm; □) was also analyzed.
The specific autoglucosylation time course of the heterodimer formed by glycogenin subunits with complementary glucosylation and acceptor capacities is shown in Fig. 5. The heterodimer formed by mixing 0.04 μm Y194F-Δ270 with 3.96 μm T82M-Δ270 (Scheme 1B) showed a lineal autoglucosylation increase without reaching a plateau (Fig. 5A). The extent of autoglucosylation at 60 min was 34 mol of glucose/mol of heterodimer. This was ∼3-fold higher than the MSAE of the wild-type dimeric enzyme and, taking into consideration the constant increase in specific autoglucosylation, may indicate a dynamic exchange of the acceptor subunit during heterodimer glucosylation, with the replacement of glucosylated by non-glucosylated T82M-Δ270. On reversing the proportions of the mutants by mixing 0.04 μm T82M-Δ270 with 3.96 μm Y194F-Δ270, a similar subunit exchange was expected to occur but, in this case, between the heterodimer and the dimer pool of the glucosylation-active Y194F-Δ270 mutant, resulting in the maximum autoglucosylation of the T82M-Δ270 heterodimer subunit. Under these conditions, the autoglucosylation was 6.3 glucose units/60 min/mol of heterodimer, reaching a plateau of 7.5 glucose units at 120 min of incubation (Fig. 5B). Thus, the rate and extent of glucopolymerization for the heterodimer were lower than for the wild-type dimeric enzyme (Fig. 2A). No autoglucosylation was measured when 0.04 μm Y194F-Δ270 and T82M-Δ270 were mixed and subjected to autoglucosylation for 60 and 120 min (Fig. 5B). This supports the existence of stable monomers at the submicromolar concentration at which the MSAE was measured, with no detectable formation of transient autoglucosylable dimers (Fig. 2).
FIGURE 5.
Time course of heterodimer intersubunit autoglucosylation. The heterodimers formed by mixing and equilibration of 0.04 μm Y194F-Δ270 with 3.96 μm T82M-Δ270 (A) and 0.04 μm T82M-Δ270 with 3.96 μm Y194F-Δ270 (●) and 0.04 μm T82M-Δ270 with 0.04 μm Y194F-Δ270 (□) (B) were subjected to 14C autoglucosylation as described under “Experimental Procedures.” [14C]Glucose incorporation is represented as picomoles of heterodimer present.
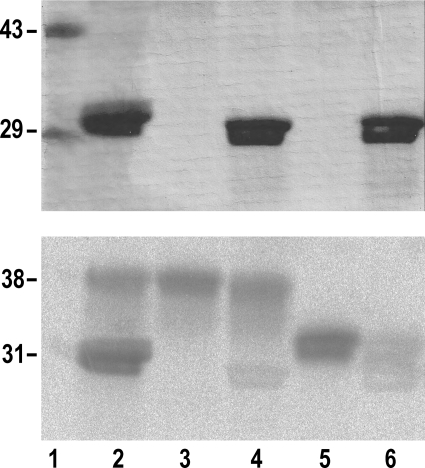
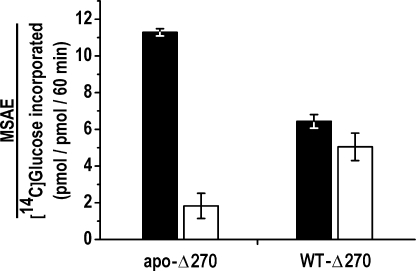
With respect to the possibility of dimer intrasubunit glucosylation, evidence that this did in fact occur was obtained by SDS-PAGE of the 14C-autoglucosylated heterodimer formed by truncated and intact glycogenin species. Simultaneous intrasubunit and intersubunit glucosylation was observed when mixing the partially glucosylated full-length WT-38 species (0.04 μm) with the T82M-Δ270 mutant (3.96 μm) having only acceptor capacity (Scheme 1E and Fig. 6, lane 2). The intrasubunit glucosylation of the WT-38 species was slightly inhibited compared with monomer autoglucosylation (Fig. 6, lane 3). A similar slight inhibition of the WT-38 intrasubunit glucosylation occurred when forming the heterodimer with the D159S/Y194F-Δ270 double mutant (Fig. 6, lane 4). On the other hand, autoglucosylation was strongly inhibited when the non-glucosylated apo-Δ270 species was subjected to dimerization with the D159S/Y194F-Δ270 mutant (Scheme 1C and Fig. 6, lanes 5 and 6). This inhibition was further examined by measuring the specific autoglucosylation extent of apo-Δ270 and WT-Δ270 after forming a heterodimer with the D159S/Y194F-Δ270 double mutant. Fig. 7 shows that the MSAE of the monomeric form was ∼5-fold lower upon heterodimerization of non-glucosylated apo-Δ270 compared with the partially glucosylated WT-Δ270 enzyme, which was only slightly inhibited, as observed in Fig. 6 for the WT-38 species.
FIGURE 6.
Simultaneous heterodimer intrasubunit and intersubunit glucosylation. The monomeric WT-38 (lanes 2–4) and apo-Δ270 (lanes 5 and 6) glycogenin species (0.04 μm) were subjected to 14C autoglucosylation for 60 min as described under “Experimental Procedures” before (lanes 3 and 5) and after mixing and equilibration with 3.96 μm dimeric T82M-Δ270 (lane 2) and D159S/Y194F-Δ270 (lanes 4 and 6), and aliquots were analyzed by SDS-PAGE. The molecular mass standards are indicated (lane 1). Upper panel, Coomassie blue stain; lower panel, autoradiogram.
FIGURE 7.
Heterodimer intrasubunit autoglucosylation. Shown is the MSAE of monomeric apo-Δ270 and WT-Δ270 glycogenins before (black bars) and after (white bars) dimerization with D159S/Y194F-Δ270. The double mutant (3.96 μm) was mixed and equilibrated with 0.03 μm apo-Δ270 or WT-Δ270 and subjected to 14C autoglucosylation as described under “Experimental Procedures.”
DISCUSSION
Two molecular forms of glycogenin are able to autoglucosylate and may thus be candidates for priming de novo biosynthesis of glycogen. An intramolecular mechanism of glucosylation has already been described for the glycogenin monomer, but the extent of the glucopolymerization capacity was not determined. On the other hand, the glycogenin dimer has been shown to synthesize an oligoglucan of 12 glucose units, but its capacity to carry this out by the proposed intersubunit glucosylation mechanism has not been demonstrated. In this work, we determined that the average MSAE produced by the glycogenin monomer is 13.3 ± 1.9 glucose units, similar to the MSAE of 12.5 ± 1.4 glucose units measured for the dimer. We also analyzed the glucopolymerization capacity of a heterodimer able to autoglucosylate only by an intersubunit or intrasubunit reaction mechanism. For this purpose, we used two single mutants, Y194F-Δ270 and T82M-Δ270, and the D159S/Y194F-Δ270 double mutant, inactive for autoglucosylation. Each of the three mutants was able to form heterodimers when mixed with apo-Δ270 (Fig. 4), and the crystal structures of the four species were indistinguishable when superimposed (Fig. 1).
As shown in Fig. 5, the time course of specific intersubunit autoglucosylation of the heterodimer formed between the Y194F-Δ270 and T82M-Δ270 mutants (Scheme 1B) revealed the incapacity of this reaction mechanism to produce the glucosylation rate or the full glucopolymerization shown by the wild-type dimeric enzyme. On the other hand, dimerization of autoglucosylation-active WT-Δ270 and apo-Δ270 with the double mutant, to analyze the glucosylation capacity of the intrasubunit mechanism (Scheme 1C), hindered the glucosylation of non-glucosylated apo-Δ270 but only slightly affected the partially glucosylated WT-Δ270 monomeric glycogenin autoglucosylation (Fig. 6). The MSAE of the monomeric enzyme forms of WT-Δ270 and apo-Δ270 was reduced by 16 and 84%, respectively, after forming a heterodimer with the D159S/Y194F-Δ270 mutant (Fig. 7). Thus, dimer intrasubunit glucosylation cannot significantly initiate but is able to complete autoglucosylation after Tyr-194 has acquired several glucose units by intersubunit glucosylation. The incapacity of the dimer to acquire the first glucose units by intrasubunit glucosylation might be due to the restrictions imposed on the dynamics of the protein, thus affecting the movement of the acceptor tyrosine toward the reaction center. The proximity of the α-helixes containing the Tyr-194 residues to the contact surface between the subunits (12) could also have produced such restriction. Once partially glucosylated by intersubunit reaction, the Tyr-bound oligoglucan is then accessible for further glucosylation by intrasubunit reaction.
The crystal structure of rabbit glycogenin in which Asp-162 was substituted with either serine or asparagine has provided evidence for the essential role of Asp-162 in the glucosylation reaction. This result led to the proposal that Asp-162 was the nucleophile involved in the formation of a covalent enzyme-substrate donor intermediate (13). Examination of the wild-type enzyme crystal structure (12) shows the carboxylate of Asp-162 to be at a distance of 21.03 Å from the Tyr-194 hydroxyl group within the same dimer subunit. A distance of 15.82 Å separates the Asp-162 carboxylate of a dimer subunit from the Tyr-194 hydroxyl group of the opposing subunit. Considering these large distances, it appears impossible for autoglucosylation to take place. On the basis of the detection of tetrameric forms of glycogenin by gel filtration, other authors have examined whether these molecular forms could allow intermolecular glucosylation of the dimer (Scheme 1F) (6, 13). However, the tetramer formation in our preparations at high enzyme concentrations was neither detected by gel filtration nor shown by analysis of the specific autoglucosylation rate dependence on protein concentration (15). It is conceivable that the molecular dynamics of the enzyme would provide the conformational modifications required to shorten the mentioned distances, allowing the glucose transfer from the glucosylated Asp-162 intermediate.
These results led us to conclude that both intersubunit glucosylation and intrasubunit glucosylation acting in combination are required to produce the full glucopolymerization of the glycogenin dimer. Nevertheless, the full autoglucopolymerization capacity of the glycogenin monomer fulfills the functional role of the enzyme to prime the biosynthesis of glycogen in vivo, immediately after the protein is expressed, without requiring prior dimerization.
Supplementary Material
Acknowledgments
We are grateful to the staff of the Laboratório Nacional de Luz Síncrotron (Campinas, Brazil) for assistance during data collection. We thank Dr. Paul Hobson for the English revision of the manuscript.
This work was supported by grants from the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), the Fondo para la Investigación Científica y Tecnológica (FONCyT), and the Secretaría de Ciencia y Técnica-Universidad Nacional de Córdoba (SECyT-UNC).

This article contains supplemental Table S1.
The atomic coordinates and structure factors (codes 3USQ and 3USR) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
Carrizo, M. E., Romero, J. M., Issoglio, F. M., and Curtino, J. A. (2012) FEBS Lett., in press.
- MSAE
- maximum specific autoglucosylation extent.
REFERENCES
- 1. Krisman C. R., Barengo R. (1975) A precursor of glycogen biosynthesis: α-1,4-glucan-protein. Eur. J. Biochem. 52, 117–123 [DOI] [PubMed] [Google Scholar]
- 2. Aon M. A., Curtino J. A. (1984) Evidence for the glycoprotein nature of retina glycogen. Eur. J. Biochem. 140, 557–566 [DOI] [PubMed] [Google Scholar]
- 3. Kennedy L. D., Kirkman B. R., Lomako J., Rodriguez I. R., Whelan W. J. (1985) in Membrane and Muscle (Berman M. C., Gevers W., Opies L. H., eds) pp. 65–84, ICSU Press/IRL Press, Oxford [Google Scholar]
- 4. Pitcher J., Smythe C., Campbell D. G., Cohen P. (1987) Identification of the 38-kDa subunit of rabbit skeletal muscle glycogen synthase as glycogenin. Eur. J. Biochem. 169, 497–502 [DOI] [PubMed] [Google Scholar]
- 5. Bazán S., Curtino J. A. (2005) The size of the C-chain maltosaccharide of glycogen: evidence for the presence of only a single branch. Glycobiology 15, 14C–18C [DOI] [PubMed] [Google Scholar]
- 6. Hurley T. D., Walls C., Bennett J. R., Roach P. J., Wang M. (2006) Direct detection of glycogenin reaction products during glycogen initiation. Biochem. Biophys. Res. Commun. 348, 374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cheng C., Mu J., Farkas I., Huang D., Goebl M. G., Roach P. J. (1995) Requirement of the self-glucosylating initiator proteins Glg1p and Glg2p for glycogen accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 15, 6632–6640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moslemi A. R., Lindberg C., Nilsson J., Tajsharghi H., Andersson B., Oldfors A. (2010) Glycogenin-1 deficiency and inactivated priming of glycogen synthesis. N. Engl. J. Med. 362, 1203–1210 [DOI] [PubMed] [Google Scholar]
- 9. Aon M. A., Curtino J. A. (1985) Protein-bound glycogen is linked to tyrosine residues. Biochem. J., 229, 269–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Smythe C., Caudwell F. B., Ferguson M., Cohen P. (1988) Isolation and structural analysis of a peptide containing the novel tyrosyl-glucose linkage in glycogenin. EMBO J. 7, 2681–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miozzo M. C., Lacoste E. R., Curtino J. A. (1989) Characterization of the proteoglycogen fraction non-extractable from retina by trichloroacetic acid. Biochem. J. 260, 287–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibbons B. J., Roach P. J., Hurley T. D. (2002) Crystal structure of the autocatalytic initiator of glycogen biosynthesis, glycogenin. J. Mol. Biol. 319, 463–477 [DOI] [PubMed] [Google Scholar]
- 13. Hurley T. D., Stout S., Miner E., Zhou J., Roach P. J. (2005) Requirements for catalysis in mammalian glycogenin. J. Biol. Chem. 280, 23892–23899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lin A., Mu J., Yang J., Roach P. J. (1999) Self-glucosylation of glycogenin, the initiator of glycogen biosynthesis, involves an intersubunit reaction. Arch. Biochem. Biophys. 363, 163–170 [DOI] [PubMed] [Google Scholar]
- 15. Bazán S., Issoglio F. M., Carrizo M. E., Curtino J. A. (2008) The intramolecular autoglucosylation of monomeric glycogenin. Biochem. Biophys. Res. Commun. 371, 328–332 [DOI] [PubMed] [Google Scholar]
- 16. Skurat A. V., Cao Y., Roach P. J. (1993) Glucose control of rabbit skeletal muscle glycogenin expressed in COS cells. J. Biol. Chem. 268, 14701–14707 [PubMed] [Google Scholar]
- 17. Leslie A. G. W. (1992) Joint CCP4/ESF-EACMB Newsletter on Protein Crystallography, No. 26, 27–33 [Google Scholar]
- 18. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 19. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Carrizo M. E., Miozzo M. C., Goldraij A., Curtino J. A. (1997) Purification of rabbit skeletal muscle proteoglycogen: studies on the glucosyltransferase activity of polysaccharide-free and -bound glycogenin. Glycobiology 7, 571–578 [DOI] [PubMed] [Google Scholar]
- 23. Kabsch W. (1976) A solution for the best rotation to relate two sets of vectors. Acta Crystallogr. A 32, 922–923 [Google Scholar]
- 24. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Developments in the CCP4 molecular graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.