FIGURE 6.
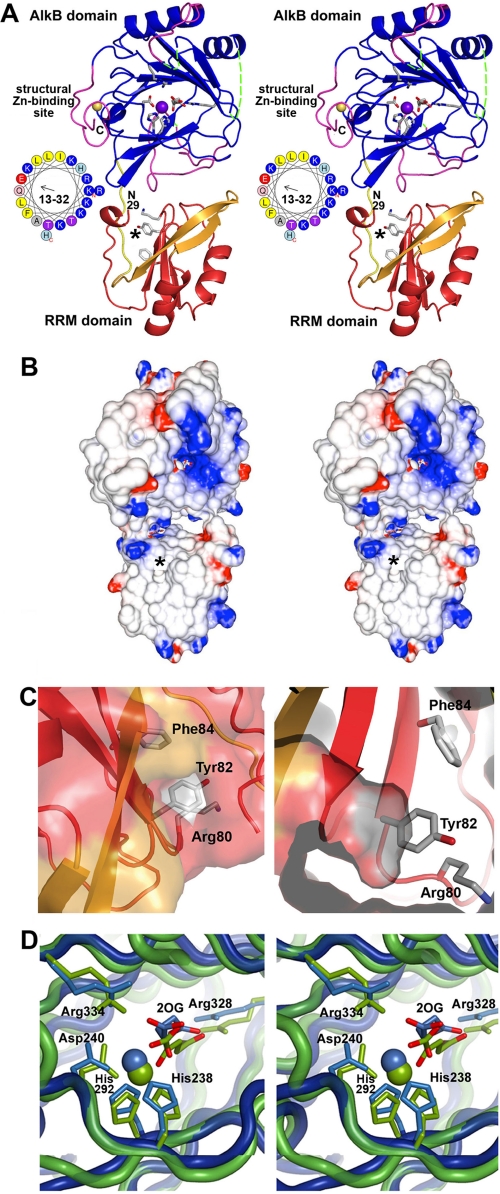
Crystal structure of the RRM/AlkB domains in human ABH8. A, stereo ribbon diagram of the structure in complex with Zn(II) and Mn(II)/2OG. The conserved and non-conserved regions in the RRM domain are colored red and orange, respectively, whereas the conserved and non-conserved regions in the AlkB domain are colored blue and magenta, respectively. The loop connecting the two domains is shown in yellow. Conserved residues in the RRM (Lys80, Tyr82, and Phe84) and AlkB (His238, Asp240, His292, Arg328, and Arg334) domains as well as the 2OG bound to the AlkB domain are shown in stick representation (with carbon and oxygen colored white and red, respectively). The Mn(II) ion bound in the active site and the Zn(II) ion bound in the C-terminal structural Zn(II)-binding site are shown as purple and yellow spheres, respectively. The asterisk in A and B marks the location of a putative pyrimidine-binding pocket in the RRM domain (shown in C). The observed N terminus of the RRM domain at residue 29 and C terminus of the AlkB domain at residue 354 are labeled in black (“N29” and “C”, respectively). Residues 13–32 in ABH8, most of which were deleted to improve crystal quality, are shown as an α-helical wheel with basic amino acids colored blue. Assessment of structural conservation and the numbering of the secondary structural elements (supplemental Fig. S7A) in the AlkB domain are based on comparison with other AlkB family enzymes (48–50) (rather than the Fe(II)/2OG dioxygenase superfamily as done in Yu et al. (35)). Residues 156–174 and 181–192 in the AlkB domain (dotted green lines), which are topologically equivalent to the segments forming the nucleotide-binding lid in E. coli AlkB, are disordered in ABH8. B, stereopair showing the electrostatic potential of the molecular surface of the domains oriented as in A. Fully saturated blue and red colors represent potentials of ±8 kT at 100 mm ionic strength as calculated by GRASP2 (60). C, two views of a putative pyrimidine-binding pocket formed in part by the RNP1 motif in the RRM domain. The entrance to this cavity, which can accommodate a pyrimidine base without steric clash, lies on the surface of the RRM domain below the active site in the AlkB domain. The entrance is marked by a black asterisk in A and B and in supplemental Fig. S7A. Residue Tyr82 is positioned at the base of the cavity (as shown in the right panel) where it could make a stacking interaction with a bound pyrimidine. The residues in the RNP1 motif are shown in stick representation colored according to atomic identity (carbon in gray, oxygen in red, and nitrogen in blue). The molecular surface is colored like the backbone except for the regions formed by the side chains of the residues in RNP1 motif, which are colored according to atomic identity. D, stereopair showing superposition of the active sites in ABH8 (blue) and E. coli AlkB (green; Protein Data Bank code 2FDH) with the invariant residues, the 2OG co-substrates, and the Mn(II) cofactors colored the same as the domains. Mn(II) is widely used in studies of Fe(II)/2OG dioxygenases as a catalytically inactive analog of Fe(II) that preserves active site stereochemistry. Although some high resolution crystal structures of Fe(II)/2OG dioxygenases have a weakly ordered H2O molecule in the final ligation position on the Fe(II), Mn(II), or Co(II) ion occupying the catalytic site, this ligation site is empty in other structures (14, 35, 48–50), as it is in the ABH8 active site shown here. However, other structures generally preserve octahedral coordination geometry for the ligating atoms as shown in D for E. coli AlkB, as opposed to the distorted geometry observed in all of the crystallographically independent views of the ABH8 active site in the structures reported in this study.