FIGURE 6.
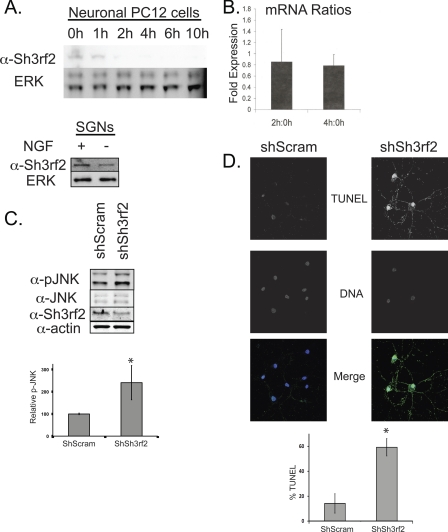
Decrease in Sh3rf2 protein is an early event in POSH-JNK-dependent apoptosis induced by trophic factor deprivation. A, top panel, PC12 cells were treated with NGF for 5–7 days and then subjected to NGF deprivation as described under “Experimental Procedures.” Cells were collected at the indicated times following treatment, and equal quantities were subjected to immunoblot as shown. Membranes were reprobed with anti-ERK2 to confirm equal loading. Bottom panel, sympathetic cervical ganglion neurons (SGNs) were treated with NGF deprivation as described under “Experimental Procedures.” Cells were collected 8 h after treatment and subjected to immunoblot as shown. B, after priming for 5 days, PC12 cells were subjected to NGF deprivation for 0, 2, or 4 h. mRNA was collected and quantitative PCR was performed as described under “Experimental Procedures.” The ratios of Sh3rf2 mRNA relative to a housekeeping gene, cyclophilin (PPIA), for each time pair were determined as indicated. C, cortical neurons were cultured for 14–21 days prior to treatment with lentivirus expressing shRNAs as indicated. 96 h later, cells were collected and subjected to immunoblot with the indicated antibodies. The same blot was reprobed serially. The Sh3rf2 band shown here runs at 55 kDa. shSh3rf2 induced a 2-fold increase of p-JNK when normalized to total JNK. The graph shows combined results from 3 experiments (*, p < 0.002 relative to control shRNA). Nearly identical results were obtained when normalizing to actin (not shown). D, murine cortical neurons were treated identically to those in C. 5 days later, cells were fixed and TUNEL-labeled as described under “Experimental Procedures.” DNA (Hoechst 33342) and TUNEL (Alexa 488) images are shown in grey scale and the merged image shown in color. Note the marked increase in TUNEL positivity and contracted, fragmented nuclei in the Sh3rf2-treated neurons. Greater than 100 cells were counted per condition. This experiment was performed twice and the graph represents the results from one experiment performed in triplicate (*, p < 0.05 versus control shRNA).