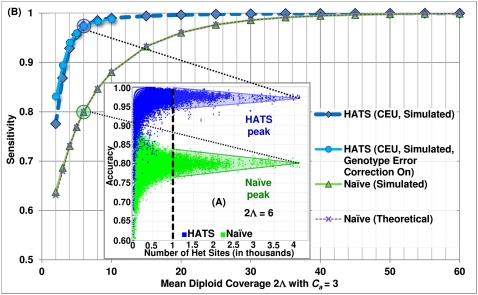
Figure 1.
Accuracy example and sensitivity of HATS and the naive method from simulations, European (CEU) training sata set. (A) Accuracies of each sample trial (2Λj = 6, Ca = 3). Each point in the embedded, raised-dot plot represents the accuracy for a particular amplicon a in sample j per trial. As the number of heterozygous sites in a increases, the accuracies converge to a peak for the naive method and a peak for HATS. We set the threshold v to 1000 and use k-means clustering to determine the centroids for each peak. The centroid for the naive model resides at ∼0.80, which is assigned as the sensitivity for the naive method for parameter values (2Λj = 6, Ca = 3). The centroid for HATS exists at ∼0.975. (B) Method sensitivities. This figure displays the simulation sensitivity results for HATS (with Genotype Error Correction [GEC] turned on or, by default, off) as well as for the naive method. The naive theoretical curve is included for comparison purposes, illustrating that the naive results can, indeed, be calculated theoretically. Note that it takes up to diploid coverage of 45 until the naive method can match the performance of HATS. The GEC mode noticeably improves performance at very low coverage levels for HATS. The training data set was obtained from the 1000 Genomes Project (http://www.1000genomes.org/).