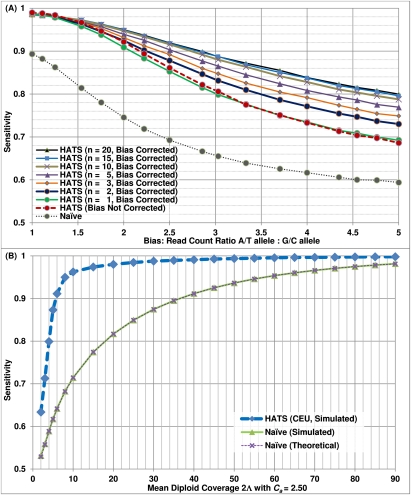
Figure 2.
Results of simulated aspects of real tumor data. (A) Sensitivity of HATS with bias correction and the naive model from simulations, European (CEU) training data set. This figure displays the simulation sensitivity results for HATS with bias correction activated (using varying sample sizes to estimate bias), bias correction inactivated, and the naive model. The x-axis represents the simulated induced bias, ranging from 1.0 (representing no bias) to 5 (representing strong bias). HATS eclipses the naive method in all instances. When the simulated bias is weak and n > 1, bias correction performs only slightly less than does no bias correction by a 0.003 cost in sensitivity. However, bias correction quickly gains the upper hand as the simulated bias increases past 1.5 (or 2 in the case of n = 2). When n = 1, the estimate of bias is not as precise and results in a slightly weaker result (with a 0.008 sensitivity cost on average) unless the induced bias is very strong (3.33). Note that there is only a marginal improvement in performance when increasing the test data set size from 15 to 20. The reason is that this reduces the training data set size, which negatively impacts sensitivity; in addition, improvement in estimation of the bias plateaus. (B) Sensitivity of HATS and the naive model from simulations with stromal contamination, European (CEU) training data set, copy number 2.5. This figure displays the simulation sensitivity results for HATS as well as for the naive model, given a copy number of 2.5, which represents a tumor of copy number 3 with 50% stromal contamination of copy-neutral healthy cells. Note that the performance gap between the two methods remains wide, and the naive method does not catch up to HATS even at a very high diploid coverage of 90. The training data set was obtained from the 1000 Genomes Project (http://www.1000genomes.org/).