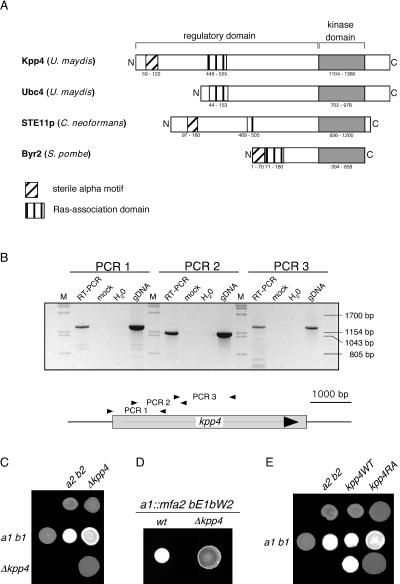
FIG. 1.
Deletion of kpp4 affects mating and filamentous growth. (A) Schematic representation of Kpp4 domain structure. Domains were identified with ISREC, and domain annotations are PS50105 (SAM domain), PS50200 (RA domain), and PS500011 (kinase domain). Accession numbers are AAN63948 for Kpp4 (1,567 amino acids), AAF86841 for Ubc4 (1,166 amino acids), AAG30205 for Ste11p of C. neoformans (1,230 amino acids), and P28829 for Bry2 of S. pombe (689 amino acids). Interestingly, at bp +4438 of the kpp4 ORF we found hexanucleotide repeats (GCTGCG) which encode an alanine stretch and are present in six copies in FB1 (a1 b1) and FB2 (a2 b2) but only in five copies in RK32 (a1 b3). (B) Reverse transcription-PCR analysis of kpp4. RNA isolated from AB33 was reverse transcribed and then subjected to three different PCRs, i.e., PCR 1 with primers kpp4-550 and OPM45, PCR 2 with primers OPM46 and OPM41, and PCR 3 with primers OPM40 and OPM21, amplifying the regions between positions −99 and +1172, +547 and +1695, and +1566 and +2872 of the kpp4 ORF, respectively. As controls, we performed reactions without reverse transcriptase (mock) or with water or genomic DNA (gDNA) as the template. Lanes M, molecular size markers. (C) The strains indicated on the top were spotted alone and in combinations with the strains indicated on the left on charcoal-containing PD plates. Dikaryotic filaments appear as white fuzziness. (D) The strains indicated the on top were spotted on charcoal-containing PD plates. SG200 developed filaments characterized by white fuzziness, while SG200Δkpp4 deletion strains are severely affected in filamentation. wt, wild type. (E) The strains indicated on the top were spotted alone (top row) and in combinations with the wild type (middle row) on charcoal-containing PD plates. In the bottom row compatible combinations of either kpp4WT (left) or kpp4RA (right) strains were cospotted.