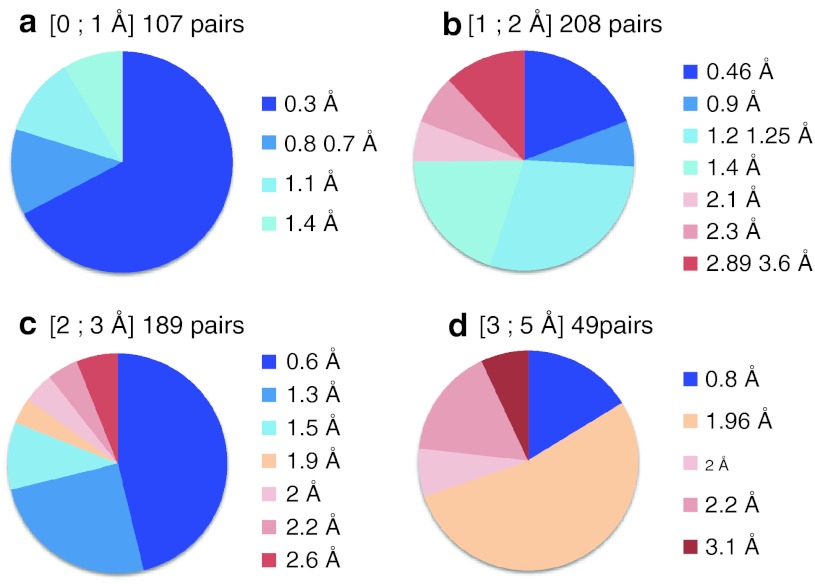
Fig. 3.
Representation of the complex pairs PKA/L1 and PKA/L2 selected by INPHARMA in dependence of the RMSD of PKA from the coordinates of the crystal structure 3DNE.pdb used as initial model for the docking (a RMSD < 1 Å; b RMSD < 2 Å; c RMSD < 3 Å; d RMSD < 5 Å). The numbers and the color code of each circle slice represent the ligands RMSD (L1, L2) from the coordinates of the ligands in the crystal structures of PKA/L1 and PKA/L2, after superimposition of the ligands. When the initial protein model is well-defined (PKA RMSD < 1 Å), INPHARMA selects complex pairs that allow ligands superposition at a resolution better than 1.5 Å. Usually, in structural based drug design, a resolution of 2 Å in the ligand coordinates is considered to be acceptable. In this limit the accuracy of the ligands superposition in the INPHARMA selected binding modes is always higher than 75%, even for ill-defined protein models (panel c and d)