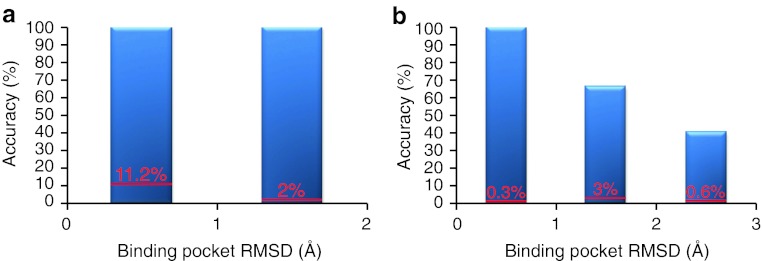
Fig. 4.
Accuracy of the INPHARMA predictions as a function of the quality of the receptor structure when the binding mode of one of the two ligands is known and used as reference. The x-axis represents the (protein only) binding pocket RMSD of the receptor models used in the docking from the crystallographic structure of PKA in the complex PKA/L1 (3DNE.pdb). The accuracy on the y axis is defined as the number of complex pairs reproducing the correct ligands superposition (absolute binding mode of L2, using the PKA/L1 complex as reference in (a); absolute binding mode of L1, using the PKA/L2 complex as reference in (b) divided by the total number of pairs selected by INPHARMA. The numbers over each bar in red represent the accuracy before applying the INPHARMA score. In this case the accuracy is the number of the complex pairs showing the correct ligands superposition divided by the total number of complex pairs selected by the energy function of the docking program. The docking for this dataset was performed with SURFLEX