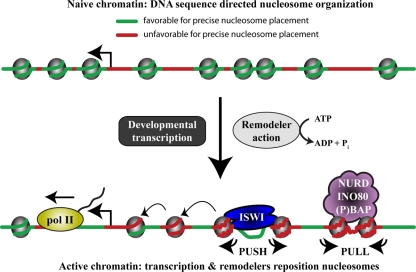
Fig 7.
Model for remodeler action. A simplified model illustrating the major conclusions from this study is shown. Replication-coupled histone deposition during early development favors DNA sequences that are characterized by strong base pair stacking energy and high bending anisotropy (green). These sequences can adopt the nucleosomal DNA trajectory, thus minimizing the entropic penalty for binding. Following the start of developmental gene expression, the combined activities of the transcription machinery and remodelers drive nucleosome repositioning. Consequently, the influence of DNA sequence properties on nucleosome organization density diminishes dramatically. The collective result of remodeler action is to increase nucleosome formation on flexible sequences with low base pair stacking energy (red). Wrapping these DNA sequences around the histone octamer requires lower deformation energy, but this would be expected to decrease the precision of nucleosome positioning and might facilitate the generation of remodeled nucleosomes. Although each remodeler class has its own characteristics, there are two main modes of chromatin remodeling. NURD, (P)BAP, and INO80 pull nucleosomes onto DNA sequences that are intrinsically unfavorable for positioning. In contrast, ISWI pushes nucleosomes away from its binding loci that innately favor nucleosome placement. Finally, remodeling can propagate from target loci and reposition neighboring nucleosomes, referred to as statistical positioning.