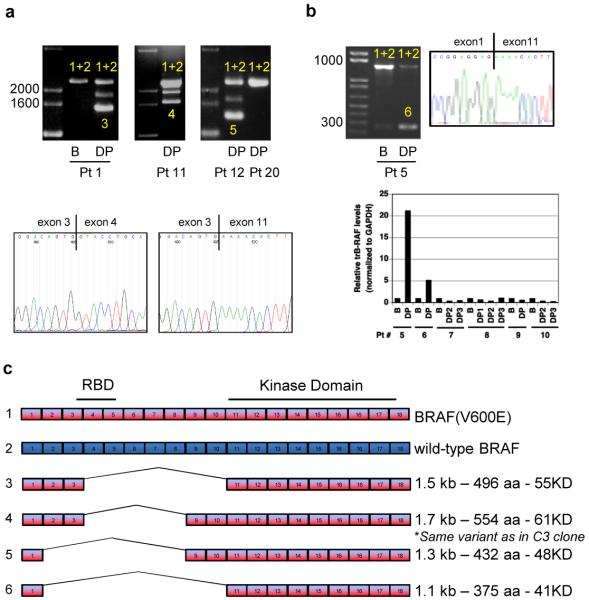
Figure 3. Identification of splice variants of BRAF(V600E) in human tumors resistant to vemurafenib.
a. PCR analysis of cDNA derived from tumor samples from patients treated with vemurafenib. In samples with only one band (full-length BRAF), both BRAF(V600E) and wild-type BRAF (1+2) were detected. In resistant tumor samples expressing shorter transcripts, the shorter transcript was a splicing variant of BRAF(V600E) (3, 4, 5). The figure shows samples from three patients with acquired resistance to PLX4032: baseline (B) and disease progression (DP) samples from patient 1 and post-treatment samples from patients 11 and 12. A tumor sample from a patient with de novo resistance to vemurafenib (patient 20) is also shown. The intermediate band in samples expressing splicing variants (Pts 1, 11, 12) is an artifact of the PCR reaction resulting from switching between two very similar templates. Representative Sanger sequencing traces showing the junction between exons 3 and 11 in the DP sample from patient 1 compared to the full-length transcript derived from the baseline pre-treatment sample. b. As in A, baseline (B) and disease progression (DP) samples from a patient with an exon 2-10 deletion. RNA/cDNA levels of the exon 2-10 deletion were determined by qPCR using an exon 1/11 junction primer. The data are shown as the average of duplicates and expressed as relative levels between patient-matched samples c. Exon organization of the splicing variants found in tumors from six patients that initially responded and then progressed on vemurafenib.