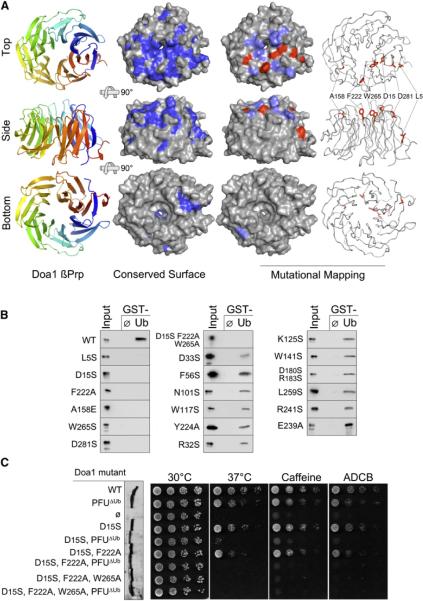
Figure 2. Structure and Function of the Ub-Binding Doa1 β-Propeller.
(A) The 1.35Å structure of the Doa1 β-Prp (residues 2–295) as viewed from the top, side, and bottom perspectives. Left column shows a cartoon of Doa1 where color spectrum of blue to red depicts N terminus to C terminus. Middle column shows the molecular surface of Doa1 β-Prp where residues conserved among Doa1 homologs are indicated in dark blue. Right columns depict summary of mutagenesis data mapped onto the Doa1 β-Prp structure. Mutations with little to no impact on Ub binding are colored light blue, and those that blocked Ub binding are colored red on the molecular surface of Doa1.
(B) Binding assays with GST alone (ø) and GST-Ub using bacterially produced wild-type and mutant Doa1 β-Prps. β-Prps were made as V5-epitope-tagged proteins and immunoblotted with anti-V5 antibodies together with a 3% equivalent of input. (C) Effect of Doa1 mutations on in vivo function. Mutant doa1Δ null cells were transformed with low-copy plasmids encoding C-terminally HA-tagged Doa1 with either no mutations (WT), a mutation that disrupts Ub binding by the central PFU domain (PFUΔUb), mutations that disrupt Ub binding by the β-Prp (residues D15, F222, W265), or a combination of UBD mutations in both the PFU and β-Prp domains. Left shows an anti-V5 immunoblot of whole-cell lysates from the corresponding transformants showing comparable expression of each Doa1-V5 mutant. Right shows growth assays of serially diluted cells grown on minimal media at 30°C, 37°C, or 30°C in the presence of 0.2% caffeine or 75 μg/ml L-azetidine-2-carboxylic acid (ADCB).