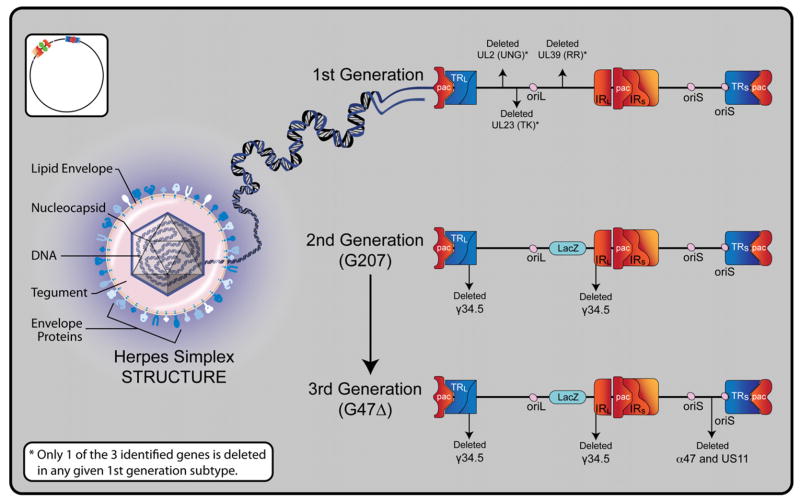
Figure 1. Overview of Oncolytic HSV-1 Structure and Generations.
Herpes simplex virus type 1 is a double-stranded DNA virus with icosahedral symmetry. A central core contains viral DNA within a nucleocapsid. Tegument between the nucleocapsid and outer envelope contains various proteins that are transferred to a host cell upon fusion. The lipid envelope carries viral glycoproteins that facilitate cellular fusion and specificity. The 152 kb DNA genome is comprised of unique long and short segments, each flanked by inverted (IRL and IRS) and terminal repeats (TRL and TRS). The genome further contains three DNA packaging (pac) signals, which enable construction of virions. There are two different origins of replication, one in the unique long segment (oriL), and one in the unique short segment (oriS). Several genes are duplicated as a result of the inverted repeats. These include oriS, γ34.5, α0, and α4. Representative constructs are shown of each of the three generations of oncolytic HSV-1. In the first generation, potential single deletions of uracil deglycosylase (UNG), ribonucleotide reductase (RR), and thymidine kinase (TK) are represented. In the second generation, the G207 construct reflects double deletions of γ34.5 and the addition of a LacZ gene at the site of UL39 (RR), which effectively inactivates RR and enables histochemical identification of viral replication via β-galactosidase detection. In the third generation, the G47Δ construct reflects its derivation from the G207 backbone with the deletion of the α47 gene and the overlapping US11 promoter region. Since α47 encodes an inhibitor of antigen presentation, its deletion prevents the down-regulation of MHC class I peptides on the surface of virally infected cells, thereby diminishing host immune responses and enhancing viral efficacy.