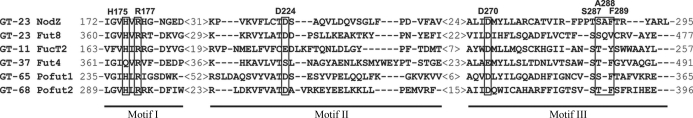
Figure 2.
Structure-based sequence alignment of the conserved sequence motifs I, II and III of α1,2-, α1,6- and protein O-fucosyltransferases. The alignment is based on the present structure of Bradyrhizobium sp. WM9 NodZ and on the following sequences retrieved from GenBank: FUT8, human α1,6-FucT (D89289); FucT2, α1,2-FucT from H. pylori (AF076779); Fut4, xyloglucan α1,2-FucT from Arabidopsis thaliana (AF417474); POFUT1, human protein O-FucT (AF375884); POFUT2, human protein O-FucT (AJ575591). Residues engaged in interactions with GDP are boxed and annotated with residue numbers in the NodZ sequence. The numbers in angle brackets indicate the number of amino-acid residues skipped in the alignment.