Table 1. Crystallographic data and refinement statistics for NodZ complexes.
Values in parentheses are for the last resolution shell.
| Data set | NodZ–GDP | NodZ–GDP-Fuc |
|---|---|---|
| Data-collection and processing statistics | ||
| Beamline | SER-CAT 22-ID | SER-CAT 22-BM |
| Wavelength (Å) | 1.0000 | 0.9724 |
| Temperature (K) | 100 | 100 |
| Space group | P6522 | P6522 |
| Unit-cell parameters (Å) | ||
| a | 123.9 | 128.8 |
| c | 95.2 | 91.1 |
| Mosaicity (°) | 0.48 | 0.70 |
| Resolution range (Å) | 50.0–1.98 (2.05–1.98) | 50.0–2.35 (2.43–2.35) |
| Total reflections | 300152 | 204336 |
| Unique reflections | 30783 | 19190 |
| Completeness (%) | 99.9 (99.9) | 99.9 (100) |
| Multiplicity | 9.8 (9.4) | 10.6 (10.1) |
| 〈I/σ(I)〉 | 16.6 (3.7) | 20.5 (3.3) |
| Rmerge† | 0.119 (0.706) | 0.091 (0.656) |
| Refinement statistics | ||
| Resolution (Å) | 50.0–1.98 | 50.0–2.35 |
| No. of reflections, working set | 29348 | 18018 |
| No. of reflections, test set | 1103 | 1030 |
| R/Rfree‡ | 0.172/0.217 | 0.210/0.258 |
| No. of atoms (protein/water) | 2333/199 | 2345/82 |
| No. of ions (phosphate/chloride) | 3/0 | 1/1 |
| R.m.s. deviation from ideal | ||
| Bond lengths (Å) | 0.019 | 0.018 |
| Bond angles (°) | 1.70 | 1.50 |
| Average B factor (Å2) | 26.7 | 47.8 |
| Ramachandran statistics (%) | ||
| Most favoured regions | 93.6 | 92.6 |
| Allowed regions | 6.4 | 7.4 |
| PDB code | 3siw | 3six |
R
merge = 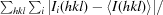
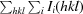 , where 〈I(hkl)〉 is the average intensity of reflection hkl.
, where 〈I(hkl)〉 is the average intensity of reflection hkl.
R = 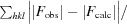
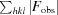 , where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is calculated analogously for the test reflections, which were randomly selected and excluded from the refinement.
, where F
obs and F
calc are the observed and calculated structure factors, respectively. R
free is calculated analogously for the test reflections, which were randomly selected and excluded from the refinement.
