Figure 6. Models for Engulfment in Protoplasts and Intact Cells.
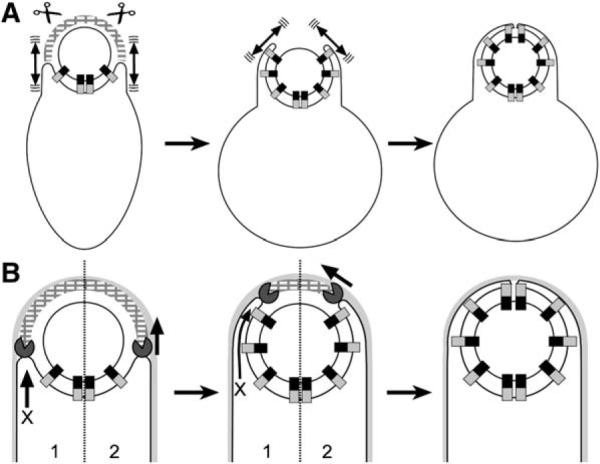
(A) Brownian ratchet model for protoplast engulfment. During protoplast formation, peptidoglycan hydrolases (scissors) remove the peptidoglycan (hatched arcs) that blocks the engulfing membrane, allowing the Q-AH ratchet to engage and prevent backward membrane movement. The mother cell membrane could move by Brownian motion (double-headed arrows).
(B) Models for engulfment in intact cells. (1) Missing motor model. The DMP proteins (Pac-Man) might act like lysozyme, degrading the peptidoglycan that blocks engulfment. Since SpoIIQ and SpoIIIAH are not essential for engulfment in whole cells, this model requires additional unidentified force-generating proteins (X arrow). (2) Dual ratchet model. The membrane-anchored DMP proteins might act as a burnt-bridge Brownian ratchet, in which the enzymatic activity of SpoIID renders complex diffusion along the peptidoglycan unidirectional, thereby moving the engulfing membrane around the forespore. The Q-AH ratchet could contribute to the directionality and processivity of the DMP ratchet.
