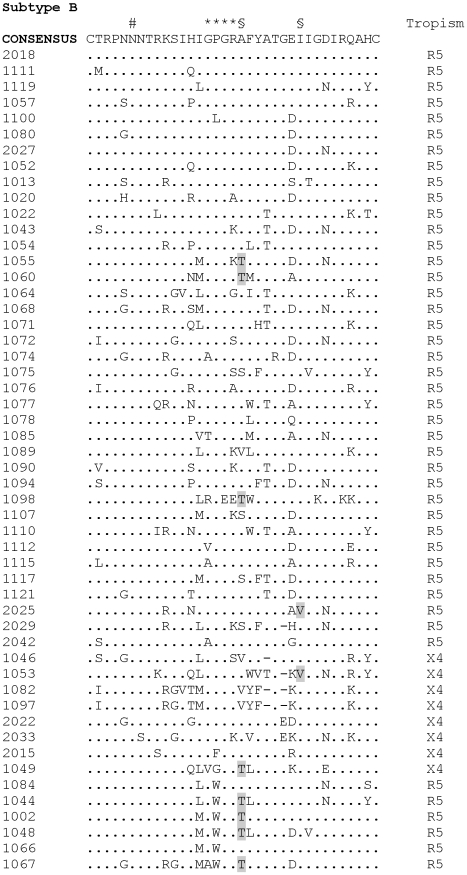
Figure 1. Amino acid alignment of V3 sequences.
Subtype B sequences were aligned with the consensus sequence. Amino acid positions appear at the top of each alignment, with dots indicating identities whereas dashes indicate deletions. # indicates the N-linked glycosylation site, * indicates the GPGR motif at the tip of the V3 loop, and § indicates positions 316 and 323 (HXB2 positions), which are associated with in vitro selection of maraviroc resistance (316T and 323 V substitutions are shaded) [29]. Inferred tropism is indicated to the right of each sequence (CXCR4-using strains are named X4 for the sake of simplicity).