Figure 4.
Structure of the CALMANTH Domain
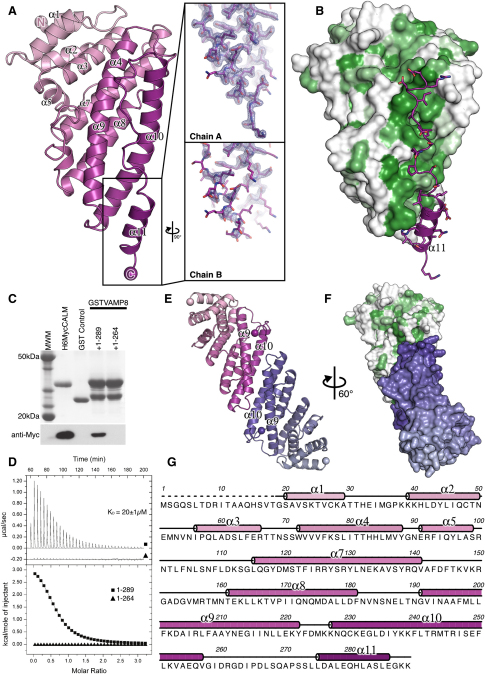
(A) Ribbon representation of the structure of the CALMANTH domain colored from pink (residue 19) to purple (residue 288). Helices are numbered as in (Ford et al., 2001) with no α6. The insets show 2FO–FC electron density contoured at 1.2 σ for the well- and poorly-ordered α11 helix in the two CALMANTH molecules in the asymmetric unit (designated chains A and B).
(B) Surface representation of helices α1-α10 of the CALMANTH domain colored from high (dark green) to low (white) hydrophobicity oriented as in (A). The hydrophobic groove in which helix α11 and the ten preceeding residues sit can be clearly seen.
(C) GST pull-down expriments using GST, GSTVAMP8, and the His6MycCALMANTH proteins indicated. Top panel: Coomassie blue stained gel. Lower panel: western blot probed with anti-myc. Residues 1–289 but not 1–264 of CALMANTH bound to GSTVAMP8.
(D) ITC quantitating the binding of residues 1–264 of CALMANTH to VAMP8. CALMANTH(1-264) (black triangles) showed no measurable binding to VAMP8 whereas WT CALMANTH(1-289) bound with a KD of 20 ± 1μM (black squares). Data for CALMANTH(1-264) is offset by −0.2 μcal/sec for clarity.
(E) Structure of the CALMANTH(19-264) dimer. One monomer is colored pink/purple and the other blue. The view is rotated by 60° around the vertical axis relative to (A).
(F) Surface representation of the CALMANTH(1-264) dimer with one monomer colored by hydrophobicity as in (B) and the other monomer colored blue, oriented as in (A). Formation of the dimer obscures the surface buried by helix α11 in CALMANTH(1-289) which is proposed to be the VAMP8 binding site.
(G) Sequence of CALMANTH(1-289). Secondary structure is shown above the sequence representing helices α1 to α11; α6 has been omitted as per (Ford et al., 2001).
See also Figure S4.