Figure 7.
Binding of CALMANTH and SNARE Complex Formation by VAMP8 Are Mutually Exclusive
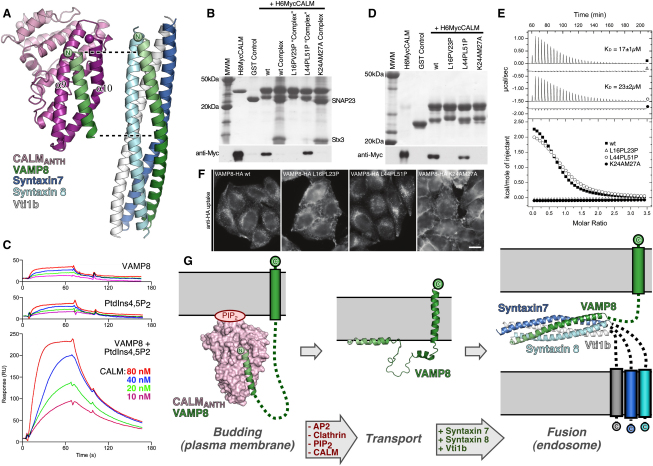
(A) Comparison of the mode of VAMP8 binding to CALMANTH and to Syntaxin7:Syntaxin8:Vti1b (PDB 1GL2) (Antonin et al., 2002) made by superimposing the VAMP8 residues 15–39 from the two complexes. VAMP8 adopts the same superhelical twist in both structures and helices α9 and α10 of CALMANTH correspond to the helices of Syntaxin 8 and Syntaxin 7, respectively.
(B) Formation of GSTVAMP8:SNAP23:Synatxin3(195–253) SNARE complexes (see Supplemental Information) and their binding to CALMANTH. Pull-down experiments using His6MycCALMANTH and the GST fusion proteins indicated. Top panel: Coomassie blue stained gel. Lower panel: western blot probed with anti-myc. SNARE complexes formed with GSTVAMP8 wt and K24AM27A but not with GSTVAMP8 L16PV23P or L44PL51P. Neither the wt VAMP8 SNARE complex nor the K24AM27A SNARE complex bound CALMANTH indicating that SNARE complex formation and CALMANTH binding shown are mutually exclusive events.
(C) Avidity of CALM binding to PtdIns4,5P2 and VAMP8. Shown are the sensorgrams for the concentration dependent binding of CALM to membranes with captured VAMP8 (top: calculated KD0.9 ± 0.15 μM), membranes with PtdIns4,5P2 (middle: calculated KD1.9 ± 0.45 μM) and membranes with both, PtdIns4,5P2 and captured VAMP8 (bottom: calculated KD0.17 ± 0.03 μM). Given are the mean values and SD of four independent measurements The affinity of CALM increases by 8.5 fold over the average KD for the two ligands when VAMP8 and PtdIns4,5P2 are bound simultaneously.
(D) Pull-down experiments using His6MycCALMANTH and wt and mutant GSTVAMP8 fusion proteins as indicated. Top panel: Coomassie blue stained gel. Lower panel: western blot probed with anti-myc. The GSTVAMP8 mutants L16PV23P and K24AM27A (see also Figure 6) do not bind CALMANTH. However, GSTVAMP8 L44PL51P bound CALMANTH with a similar strength to wt GSTVAMP8.
(E) ITC quantitating the binding of point mutated versions of VAMP8 to CALMANTH. The binding of wt CALMANTH and GSTVAMP8 L44PL51P (open circles) was comparable to that of wt CALMANTH and wt GSTVAMP8 (black squares) (KDs of 23 ± 2μM and 17 ± 1μM respectively). However, GSTVAMP8 L16PV23P (open triangles) and GSTVAMP8 K24AM27A (black circles) both showed no measurable interaction with CALMANTH. Data for GSTVAMP8 L16PL23P, L44PL51P and K24AM27A are translated by −0.3 μcal/s, −1.5 μcal/s and −1.9 μcal/s respectively for clarity.
(F) Localization of anti-HA in cells expressing different VAMP8-HA constructs. The cells were allowed to endocytose the antibody for 40 min, then processed for immunofluorescence. The cells expressing wt VAMP8 and the L44PL51P mutant have endocytosed the antibody, but the cells expressing the L16PV23P mutant have mainly retained the antibody on the plasma membrane similar to the K24AM27A mutant. The scale bar represents 20 μm.
(G) Schematic representation of the model of VAMP8 trafficking from the plasma membrane. The clathrin adaptor CALM binds simultaneously to the R-SNARE VAMP8 and PtdIns4,5P2 (labeled PIP2) at the plasma membrane. CALM is released from the surface of an endocytosed vesicle when PtdIns4,5P2 hydrolyzed and the clathrin cage disassembled and the hydrophobic CALM-binding helix of VAMP8 now “lies” on the vesicle's surface (Ellena et al., 2009). Finally VAMP8 forms a trans-SNARE complex with its cognate SNAREs on an early endosome to drive vesicule fusion. Thus throughout the interaction of the hydrophobic VAMP8 SNARE motif with the aqueous environment is minimized.