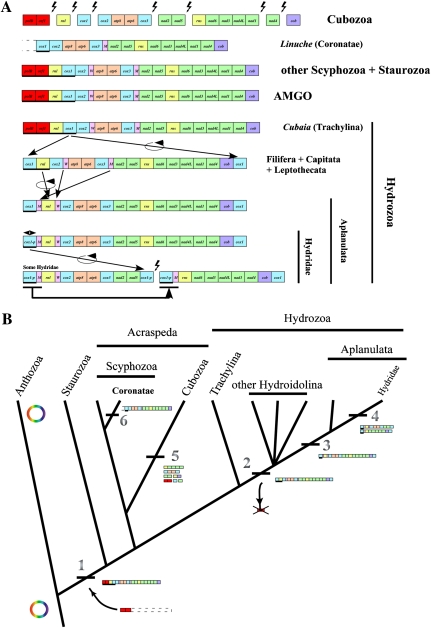
FIG. 2.—
Evolution of mitochondrial genomes in Medusozoa (Cnidaria) based on phylogenetic relationships according to Collins et al. (2006), Collins et al. (2008), and Cartwright et al. (2008). (A) Comparison of mitochondrial gene orders between various medusozoan clades and the putative mitochondrial AMGO. Breaks in contigs mark the ends of linear mitochondrial chromosomes. Genes are transcribed from left to right unless underlined. cox1-p corresponds to the partial copy of cox1 at the end of the mtDNA molecule(s) in the Hydridae family. polB is an inferred member of the B DNA polymerase gene family and ORF314 is an unidentified ORF located upstream of it. Lightning bolts represent breaks in the mitochondrial chromosomes. Black arrows depict directions of inferred genome rearrangements. (B) Evolution of genome organization of the linear mtDNA in Medusozoa. The phylogeny is based on Collins et al. (2006). Major genomic rearrangements are labeled. 1) Linearization of the mtDNA by insertion of a linear plasmid of unknown source; 2) loss of polB and ORF314, and duplication of cox1 at each end of the linear chromosome within Hydrozoa after the divergence of Trachylina; 3) displacement of tRNAs and inversion of rnl in Aplanulata; 4) genome segmentalization of the mtDNA in several Hydridae species with consequent trnM and cox1 duplication; 5) high level segmentalization of the mtDNA in cubozoan; and 6) displacement of trnW in the coronate L. unguiculata.