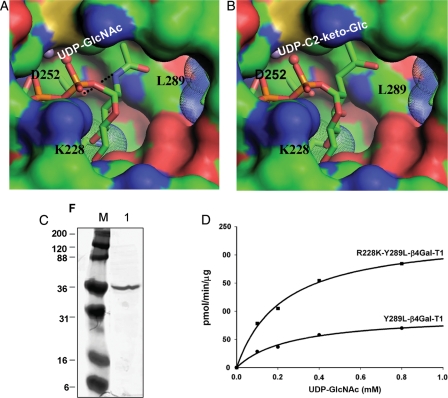
Fig. 1.
Modeling of UDP-GlcNAc (A) and UDP-C2-keto-Glc (B) binding to mutant R228K-Y289L- β4Gal-T1 molecule. The modeling was based on the UDP-GalNAc and UDP-Glc binding to the wild-type β4Gal-T1 molecule (PDB 1OQM and 1O23, respectively) and the structure of the R228K-β4Gal-T1 molecule (PDB 1YRO). The hydrogen bond between the amino group of the donor GlcNAc molecule and the side-chain carboxylate oxygen atom of the Asp252 residue is shown in a black dotted line in (A), and this interaction is absent in (B). The modeling was performed without any energy minimization and the C2-keto-Glc geometry was generated by ChemSketch program. (C) PAGE showing the purified R228K-Y289L-β4Gal-T1 protein after affinity column purification. (D) The catalytic activity of Y289L-β4Gal-T1 and the R228K-Y289L-β4Gal-T1 enzymes measured at 37°C for 10 min, using UDP-GlcNAc as the donor substrate with 25 mM β-benzyl-GlcNAc as the acceptor substrate.