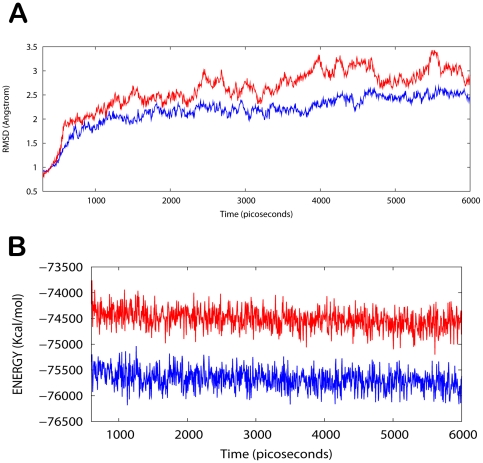
Figure 5. Analysis of MD trajectories.
(A) Plot of root mean square deviation (RMSD) of Cα of TPX2-Aurora A (protein) and TPX2/Aurora A /withanone (complex). RMSDs were calculated using the initial structures as templates. For protein (red) the reference is the PDB structure and for complex (blue) the reference is the initial docked structure. The trajectories were captured every 2.5 ps until the simulation time reached 6000 ps. (B) Plot of total energy of TPX2-Aurora A and TPX2/Aurora A/withanone (complex). The energy trajectories of both the protein (red) and the complex (blue) are stable over the entire length of simulation time with the energy of the complex always lower than that of the protein.