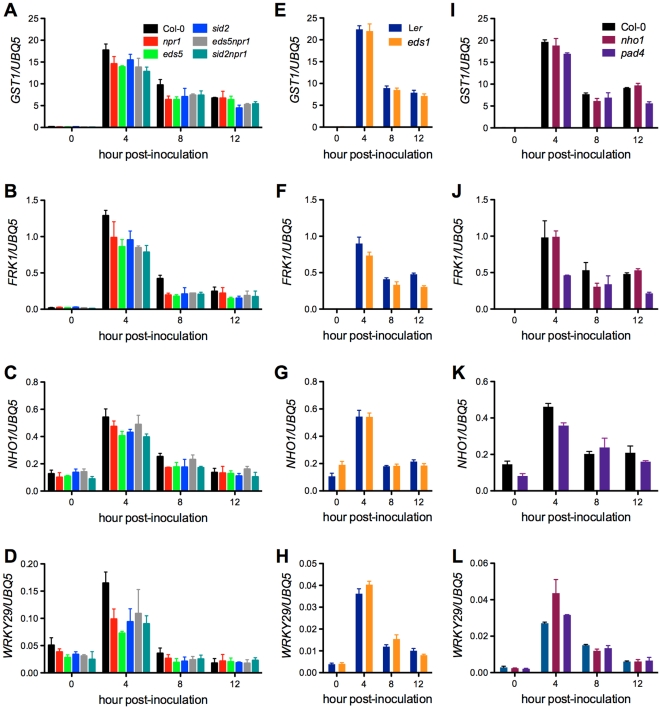
Figure 8. Expression of early response genes in the Xcc susceptible mutants.
(A to D) Expression of GST1, FRK1, NHO1, and WRKY29 in npr1, eds5, sid2, eds5npr1, and sid2npr1. (E to H) Expression of GST1, FRK1, NHO1, and WRKY29 in eds1. (I to L) Expression of GST1, FRK1, NHO1, and WRKY29 in nho1 and pad4. Four-week-old plants were inoculated with Xcc (OD600 = 0.02). Leaf tissues were collected at different time points (0, 4, 8, and 12 hpi) for total RNA isolation and gene expression analysis using RT-qPCR. Expression levels were normalized against constitutively expressed UBQ5. Data represent the mean of three biological replicates with standard deviation. Mutant eds1 is in Ler genetic background, whereas others (nho1, eds5, pad4, sid2, npr1, eds5npr1, and sid2npr1) are in Col-0 genetic background. The experiment was repeated twice with similar results.