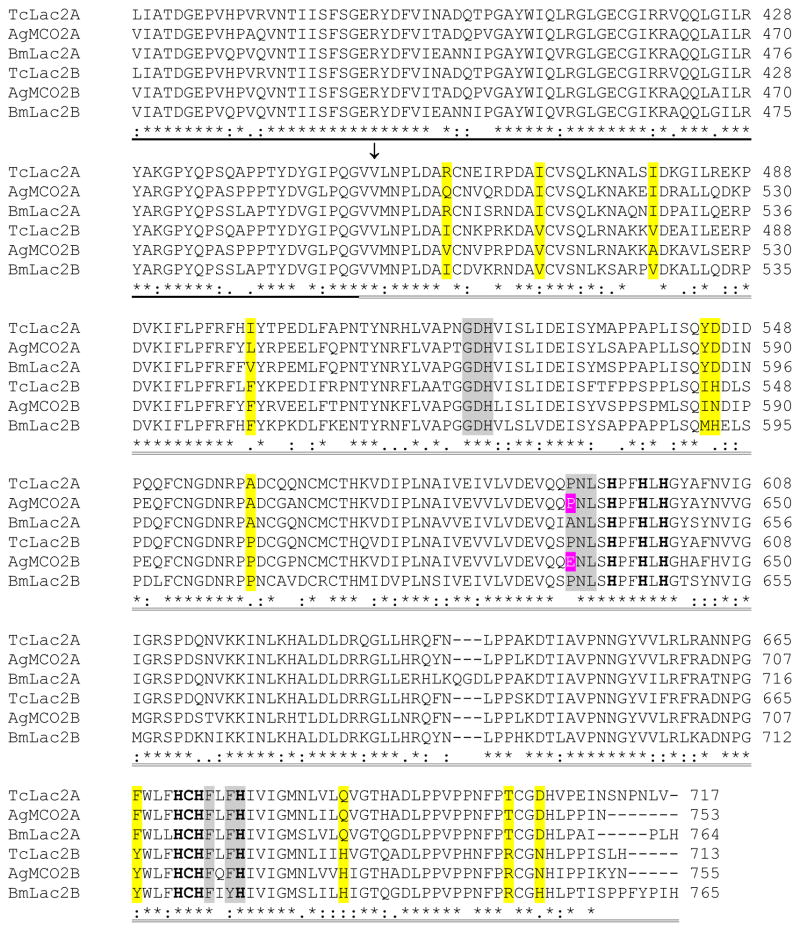
Figure 1.
Alignment of the predicted amino acid sequences of laccase-2 isoforms from T. castaneum, A. gambiae, and B. mori. Predicted signal peptides are in italicized text. Amino-terminal sequences of purified recombinant enzymes are highlighted in black. A conserved cysteine-rich region is underlined. The three cupredoxin-like domains are indicated by dashed underlining (I), bold underlining (II), and double underlining (III). (The putative boundaries of the cupredoxin-like domains were based on the crystal structure of TvLacIIIb [Bertrand et al., 2002]). An arrow points to the first residue of the alternatively spliced exons. The 10 histidines and 1 cysteine that are predicted to bind copper are in bold text. Conserved differences between the A and B isoforms are highlighted in yellow. Residues in domain III that are predicted to be part of the substrate binding pocket are highlighted in magenta (residue 633 in AgMCO2) or gray.