Figure 4.
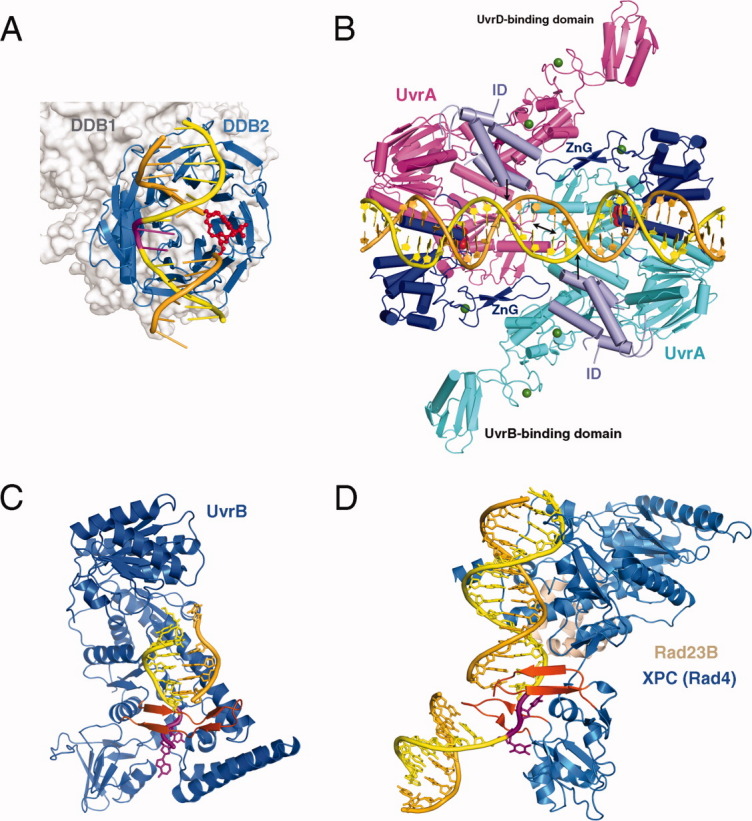
Proteins involved in NER. A: XPE complexed with 6-4 PP (PDB: 3EI1). The 6-4 PP is highlighted in red and the bases opposite it shown in magenta. DDB2 is a β-propeller protein and shown as blue ribbons. DDB1 binds DDB2 only and is shown in silver molecular surface in the background. B: UvrA-DNA complex (PDB: 3PIH). The bulk of two UvrA subunits are shown in cyan and pink ribbons diagrams except for the ATPase domains that directly contact DNA and contain the C-terminal Zinc-finger domain (dark blue) and the ID domains (pale blue). The UvrB-binding domains are labeled. Pyrophosphates marking the nucleotide-binding site are shown as red spheres, and Zn2+ as green spheres. The unstacked central G:C base pairs are indicated by a black double-headed arrow and the location of fluorescein labeled bases are marked by black arrows. C: UvrB – DNA complex (PDB: 2FDC). The protein is shown as a blue ribbon diagram, and the β hairpin that threads through DNA duplex and clamps down one strand is highlighted in orange-red. The flipped-out bases are highlighted in magenta. They are inserted into a conserved binding pocket. D: The structure of yeast homolog of XPC (Rad4) bound to a UV-damaged DNA. XPC is shown in blue and the partner protein Rad23 in light orange. Two β hairpins (in orange red) “hug” the un-damaged strand, while the damaged bases are flipped out and disordered.
