Abstract
Guanosine triphosphatases (GTPases) comprise a superfamily of proteins that provide molecular switches to regulate numerous cellular processes. The “GTPase switch” paradigm, in which a GTPase acts as a bimodal switch that is turned “on” and “off” by external regulatory factors, has been used to interpret the regulatory mechanism of many GTPases. Recent work on a pair of GTPases in the signal recognition particle (SRP) pathway has revealed a distinct mode of GTPase regulation. Instead of the classical GTPase switch, the two GTPases in the SRP and SRP receptor undergo a series of conformational changes during their dimerization and reciprocal activation. Each conformational rearrangement provides a point at which these GTPases can communicate with and respond to their upstream and downstream biological cues, thus ensuring the spatial and temporal precision of all the molecular events in the SRP pathway. We suggest that the SRP and SRP receptor represent an emerging class of “multistate” regulatory GTPases uniquely suited to provide exquisite control over complex cellular pathways that require multiple molecular events to occur in a highly coordinated fashion.
Keywords: GTPases, molecular recognition and regulation, signal recognition particle, protein targeting and translocation, protein interaction dynamics
Introduction
It is a great honor to receive the Irving Sigal Young Investigator Award from the Protein Society, and truly humbling to be added to the list of brilliant protein scientists who have previously received this award. I only hope that this serves as a motivation to set higher standards for ourselves and to carry out science at the level that deserves to be in the company of these great scientists.
I had the great fortune of being mentored by some of the most brilliant scientists. My undergraduate advisor Richard N. Armstrong and graduate advisor Daniel Herschlag taught me the skills of mechanistic enzymology—the ability to define the thermodynamic and kinetic parameters of a reaction and to translate this quantitative information into molecular mechanisms. My postdoctoral mentor Peter Walter exposed me to many exciting advances in cell biology and guided me to frame my mechanistic explorations in ways that address important biological questions. The combination of this training defined my general research goal: to understand complex cellular processes at the level of quantitative energetic principles and molecular mechanisms. I am fascinated by the following questions: how do cells build molecular switches to regulate complex pathways? What provides the driving force, and what ensures the fidelity of these processes? What properties of biological macromolecules enable them to provide such exquisite regulation?
Signal Recognition Particle (SRP) and SRP Receptor (SR): An Exception to the “GTPase Switch” Paradigm
In choosing a project for my independent research, my attention was drawn to the GTPase superfamily of proteins, which provide molecular switches to regulate numerous cellular processes. For many signaling GTPases, pioneering work has established a GTPase switch paradigm,1 in which a GTPase relies on the recruitment of external factors, such as guanine nucleotide exchange factors (GEFs) and GTPase activating proteins (GAPs), to switch between a guanosine diphosphate (GDP)-bound inactive conformation and a guanosine triphosphate (GTP)-bound active conformation [Fig. 1(A)]. A pair of GTPases in the SRP and SR stood out as notable exceptions to this paradigm. These GTPases by themselves exhibit no significant conformational changes between the apo-, GDP-, and GTP-bound states2 and exhibit rapid nucleotide exchange rates3–5 [Fig. 1(B), steps 1(a,b)]. Thus there is no need to recruit an external GEF to facilitate GDP → GTP exchange and turn these GTPases to the “on” state. Another distinguishing feature of the SRP and SR is that they form a stable heterodimeric complex in which they reciprocally activate each other's GTPase activities5,6 [Fig. 1(B), steps 2,3]. Thus there is also no need to recruit an external GAP to facilitate GTP hydrolysis and turn these GTPases to the “off” state. Indeed, SRP and SR represent a growing number of GTPases that do not adhere to the classical GTPase switch paradigm, but are instead regulated by nucleotide-dependent dimerization.7 Members of this family regulate a variety of cellular processes, including protein targeting and translocation, antiviral responses, metal ion metabolism, tRNA editing, cytoskeletal organization, endocytosis, and mitochondrial dynamics.7 This raises an important question: how do these very unusual GTPases, which lack a classical GTPase switch and do not require external GEFs or GAPs, act as molecular switches to regulate complex cell biology? The SRP and SR have provided an excellent model system to address this question; in turn, exploring this question in the SRP pathway has provided a deeper understanding of the molecular mechanisms that underlie this essential cellular pathway.
Figure 1.
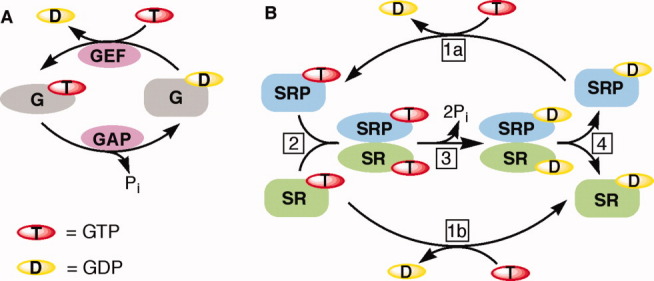
Comparison between the classic GTPase switch and the SRP and SR GTPases. (A) The bimodal GTPase cycles of classical signaling GTPases. GEF, GAP. (B) The nucleotide-driven dimerization cycle of the SRP and SR GTPases. Steps 1a and 1b, nucleotide exchange on SRP and SR, respectively. Step 2, complex formation between the SRP and SR GTPases. Step 3, activated GTP hydrolysis from the SRP•SR complex. Step 4, dissociation of SRP and SR after GTP hydrolysis, returning these GTPases to the basal state.
SRP: A Universally Conserved Protein Targeting Machine
SRP is a key cellular machinery that couples the ongoing synthesis of proteins to their proper localization, and has often served as a paradigm for understanding the molecular basis of protein localization within the cell.8,9 As in many complex cellular processes, the SRP pathway involves a series of highly orchestrated molecular steps that include: (i) recognition of nascent proteins emerging from a translating ribosome based on “signal sequences” that define their cellular destination [Fig. 2(B), step 1]; (ii) delivery of the ribosome-nascent chain complex (referred to as RNC or cargo) to the correct membrane via the interaction of the SRP with the SR (Fig. 2, steps 2,3); (iii) Unloading of the cargo to the Sec61p translocation machinery, which translocates the growing polypeptide across the membrane or integrates it into the membrane bilayer [Fig. 2, steps 4,5]; and (iv) disassembly and recycling of the SRP and SR for subsequent rounds of targeting [Fig. 2, step 5]. Each of these molecular steps requires exquisite spatial and temporal control, and my goal was to decipher how such regulation is accomplished by the highly unusual GTPase cycle of the SRP and SR.
Figure 2.
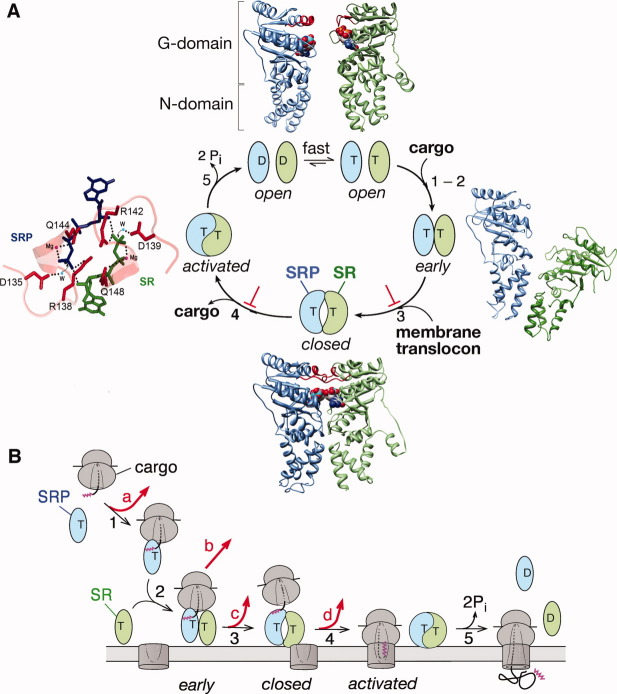
Conformational dynamics of the SRP and SR GTPases drive and regulate protein targeting. All the steps are numbered to be consistent between parts (A) and (B). The SRP GTPase is in blue, the SR GTPase is in green. T and D denote GTP and GDP, respectively. (A) Conformational changes during the SRP-SR interaction and their allosteric regulation by biological cues. The catalytic loops are highlighted in red, and the GTP analogues are in spacefill. “⊥'” denotes the pausing effect from cargo. Top panel: the crystal structures of the free SRP and SR GTPases from T. aquaticus (1JPJ and 2Q9B, respectively). “G” and “N” denote the two subdomains in SRP and SR's GTPase module. Right panel: a molecular model of the early intermediate (2XKV). Bottom panel: Cocrystal structure of the T. aquaticus SRP-SR GTPase complex (1RJ9) in the closed/activated state. Left panel: Active site interactions mediated by the catalytic loops. The GMPPCP molecules from SRP and SR are in blue and green, respectively, active site Mg2+ are in magenta, nucleophilic waters (W) are in blue, and the side chains of catalytic residues are in red. (B) GTPase rearrangements provide multiple regulatory points during protein targeting, as described in the text. At each step, the cargo can be either retained in (black arrows) or rejected from (red arrows) the SRP pathway.
What Drives SRP and SR's GTPase Cycle?
Taking an enzymologist's approach, I first set out to establish a mechanistic framework for how these two GTPases bind and reciprocally activate one another. This seemingly simple project yielded many surprises. The combination of molecular genetics, fluorescence spectroscopy, and structural analyses showed that the interaction between SRP and SR is a highly dynamic process that involves an elaborate series of discrete conformational changes. Both GTPases by themselves are in an inactive “open” conformation suboptimal for binding one another [Fig. 2(A), top structures].10,11 Their complex assembly begins with the formation of a transient early intermediate, which is driven by electrostatic attractions between sites quite distinct from the final interaction surface [Fig. 2(A), right structure].12–14 Extensive rearrangements in both GTPases then generate a more stable closed complex, in which the GTPase-domains form close contacts with one another and the two GTP molecules directly interact across the dimer interface [Fig. 2(A), bottom structure].15–17 Finally, a cooperative rearrangement of the catalytic loops in both proteins activates both GTPase sites [Fig. 2(A), left structure],15–17 and GTP hydrolysis drives the disassembly and recycling of the complex [Fig. 2(A), step 5]. Thus unlike the bimodal GTPase switch, the GTPase cycle of SRP and SR is driven by multiple conformational transitions during their dimerization that culminates in the reciprocal activation of GTP hydrolysis in their complex.
From a biophysical perspective, the SRP and SR have turned out to be an excellent system to study the dynamics of protein interactions: fluorescent dyes can be developed that detect and distinguish between different conformational states, and mutant GTPases and nucleotide analogues can be obtained that block the conformational changes at specific stages.13,17,18 This was an additional bonus offered by the SRP system that I have not anticipated in the beginning, and the potential of this system in helping us to understand macromolecular interaction dynamics and the nature and structure of transient intermediates during these interactions14 remains to be fully explored.
How Do GTPase Rearrangements Regulate Protein Targeting?
The more challenging task then was to understand whether and how the fascinating conformational dynamics in the SRP•SR complex is coupled to their biological function. Instrumental to this effort has been our collaboration with Christiane Schaffitzel and Nenad Ban's groups, who helped us set up an efficient system to generate and purify large quantities of RNCs, and who subsequently carried out beautiful structural studies that complemented our biochemical analyses (see later). In addition, my colleagues Douglas Rees and Bil Clemons taught us how to work with membrane vesicles and the translocation machinery, and I had the good fortune of having a group of brilliant graduate students and postdoctoral fellows who were unafraid to work with these large complexes. Our efforts were rewarded with many intriguing and unexpected findings, which together showed that each of the conformational changes during the binding and activation of SRP and SR provides an important regulatory point at which they can sense and respond to their biological cues and thus ensure the temporal and spatial precision of different molecular steps during the protein targeting reaction. A few of the highlights are described below.
A long-standing puzzle in the studies of the SRP-SR interaction was that although these proteins can form a stable complex, in the absence of biological cues their interaction is extremely slow (kon ∼ 102 M−1 s−1) and insufficient to sustain their cellular function. Resolution of this problem came from the finding that assembly of a stable SRP-SR complex is accelerated over 103-fold by the cargo,18–20 and 160-fold by phospholipid membranes.21 Thus the targeting reaction is a kinetically controlled process: the SRP and SR only form a complex at a physiologically relevant rate when each of them is presented with its proper biological cue. These rate accelerations ensure rapid delivery of cargo to the target membrane, and avoid futile interactions between the free SRP and SR [Fig. 2(B), step 2].
As a protein targeting machine, SRP needs to bind tightly to its cargo at the beginning of the targeting reaction [Fig. 2(B), step 1] and then at the membrane, switch to a cargo-releasing mode to unload its cargo onto the translocation machinery [Fig. 2(B), steps 3,4]. The tight binding of SRP with the RNC, though beneficial at early stages, poses a serious problem at this stage. An elegant solution to this problem is provided by the conformational changes of the GTPase complex from the early intermediate to the closed and activated states, which weakens the affinity of SRP for the translating ribosome ∼ 400-fold18 and provides an essential driving force for the handover of cargo.22 A series of structural analyses by our collaborators further provided valuable insights into how these GTPase rearrangements drive the handover of cargo. In both the RNC•SRP and RNC•SRP•SR early complex, the GTPase domains of the SRP (and SR) are localized at the nascent peptide exit site, which would block the release of the translating ribosome and shield the translocation machinery from docking onto the ribosome.12,23,24 Intriguingly, the early → activated rearrangement in the GTPase complex appears to triggered a global reorganization of the entire SRP, relocalizing the SRP•SR GTPase complex 97 Å away from its initial docking site in the SRP.25 Modeling the ribosome onto this structure suggested that such a movement would detach the SRP•SR GTPase complex from the ribosome exit site, enabling this site to form initial contacts with the Sec61p translocation machinery and thus initiate protein translocation.25 This attractive model remains to be tested.
The most intriguing effect of the cargo is “pausing,” i.e., its ability to selectively stabilize the SRP•SR complex in the early conformation and delay GTPase activation [Fig. 2(A), “⊥”].18 One possible role of pausing is that it could provide an important spatial checkpoint to prevent premature GTP hydrolysis, giving the targeting complex an important time window to search for the membrane translocon [Fig. 2(B), steps 4 vs. 5]. Consistent with this model, the interactions of SR with the target membrane relieve the cargo induced pausing and drive the rearrangements to the closed and activated states [Fig. 2, steps 3,4],21 from which the targeting reaction can be efficiently completed. Likewise, formation of an active SRP•SR complex exposes SR's lipid binding helix and allows the targeting complex to stably associate with the membrane.21 These allosteric communications allow the membrane delivery of cargo to be efficiently coupled to their subsequent unloading and translocation, thus providing exquisite spatial coordination during protein targeting. We speculate that similar roles could be played by the translocation machinery, and this hypothesis remains to be directly tested. As described below, pausing also provides an important fidelity checkpoint that helps the SRP reject the incorrect cargos.
How Is Fidelity Achieved During Protein Localization?
Signal sequences, which determine protein localization, are highly divergent and lack a consensus motif.26–28 How cellular targeting machineries select their correct substrates based on such degenerate “signals” has been elusive. Our ability to quantitatively dissect the individual molecular steps in the SRP pathway also provided us with the necessary tools to address this fundamental question. Although much of the previous work has focused on the inability of SRP to bind strongly to “incorrect” cargos [Fig. 2(B), red arrow a], we found that the initial cargo binding step is not sufficient to reject all the incorrect cargos.20 Instead, the ability of SRP and SR to undergo multiple conformational changes provides important fidelity checkpoints to help reject the incorrect cargos [Fig. 2(B), red arrows].20 For example, incorrect cargos are delivered to the membrane 3000-fold slower than the correct cargos (red arrows b-c); they are also more likely to be rejected through premature GTP hydrolysis (red arrow d). A mathematical analysis showed that all of these checkpoints are required to reproduce the experimentally determined pattern of substrate selection by the SRP.20 These results illustrate that the SRP pathway achieves high fidelity through a variety of mechanisms including binding, induced fit, and kinetic proofreading. These principles resonate with findings in the DNA and RNA polymerases and tRNA selection by the ribosome, and may represent a general mechanism by which complex cellular pathways achieve high fidelity.
Perspective
In summary, the SRP has provided an excellent model system to decipher a complex cellular process at the level of quantitative energetic principles and molecular mechanisms, and to elucidate the mode of regulation of a distinct class of regulatory GTPases. Our findings led us to propose that, in contrast to the classic bimodal GTPase switch, the SRP and SR represent an emerging class of “multistate” regulators that have the intrinsic ability to undergo conformational changes driven by distinct biological cues. A distinguishing feature of these GTPases is that it is difficult to define a single on or off state for them [Fig. 1(B)], and the biological events they mediate generally involve a complex series of molecular interactions where different functions must be turned on or off at appropriate stages of the pathway. The ability of these GTPases to undergo multiple and discrete conformational changes is critical for their role in driving cyclic processes where multiple factors must bind and later dissociate in a sequential and highly coordinated manner. A second distinguishing feature of these GTPases is their ability to intrinsically regulate their own catalytic activities without the need to recruit additional factors, which may be especially beneficial for vectorial processes that must occur quickly while maintaining high fidelity. Extensive work will be needed to decipher the precise roles and molecular mechanisms of these GTPases and to explore the extent to which multistate regulatory GTPases are involved in coordinating other important cellular processes. I hope that our work paved some of the first steps towards this goal, and that the tools and approaches we have developed provide generally applicable methodologies to help dissect other complex cellular processes.
Acknowledgments
We thank Shen K for comments on the manuscript. This work was supported by NIH grant GM078024, and career awards from the Burroughs Welcome Foundation, the Henry and Camille Dreyfus foundation, the Arnold and Mabel Beckman foundation, and the David and Lucile Packard foundation to S.S. D. A. was supported by NIH/NRSA training grant 5T32GM07616.
References
- 1.Bourne HR, Sanders DA, McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- 2.Freymann DM, Keenan RJ, Stroud RM, Walter P. Structure of the conserved GTPase domain of the signal recognition particle. Nature. 1997;385:361–364. doi: 10.1038/385361a0. [DOI] [PubMed] [Google Scholar]
- 3.Jagath JR, Rodnina MV, Lentzen G, Wintermeyer W. Interaction of guanine nucleotides with the signal recognition particle from Escherichia coli. Biochemistry. 1998;37:15408–15413. doi: 10.1021/bi981523a. [DOI] [PubMed] [Google Scholar]
- 4.Jagath JR, Rodnina MV, Wintermeyer W. Conformational changes in the bacterial SRP receptor Ftsy upon binding of guanine nucleotides and SRP. J Mol Biol. 2000;295:745–753. doi: 10.1006/jmbi.1999.3427. [DOI] [PubMed] [Google Scholar]
- 5.Peluso P, Shan S, Nock S, Herschlag D, Walter P. Role of SRP RNA in the GTPase cycles of Ffh and Ftsy. Biochemistry. 2001;40:15224–15233. doi: 10.1021/bi011639y. [DOI] [PubMed] [Google Scholar]
- 6.Powers T, Walter P. Reciprocal stimulation of GTP hydrolysis by two directly interacting GTPases. Science. 1995;269:1422–1424. doi: 10.1126/science.7660124. [DOI] [PubMed] [Google Scholar]
- 7.Gasper R, Meyer S, Gotthardt K, Sirajuddin M, Wittinghofer A. It takes two to tango: regulation of G proteins by dimerization. Nature Rev Mol Cell Biol. 2009;10:423–429. doi: 10.1038/nrm2689. [DOI] [PubMed] [Google Scholar]
- 8.Pool MR. Signal recognition particles in chloroplasts, bacteria, yeast and mammals. Mol Membr Biol. 2005;22:3–15. doi: 10.1080/09687860400026348. [DOI] [PubMed] [Google Scholar]
- 9.Miller JD, Bernstein HD, Walter P. Interaction of E. coli fFh/4.5S ribonucleoprotein and Ftsy mimics that of mammalian signal recognition particle and its receptor. Nature. 1994;367:657–659. doi: 10.1038/367657a0. [DOI] [PubMed] [Google Scholar]
- 10.Padmanabhan W, Freymann DM. The conformation of bound GMPPNP suggests a mechanism for gating the active site of the SRP GTPase site. Structure. 2001;9:859–863. doi: 10.1016/s0969-2126(01)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reyes CL, Rutenber E, Walter P, Stroud RM. X-ray structures of the signal recognition particle receptor reveal targeting cycle intermediates. PLoS One. 2007;2:e607. doi: 10.1371/journal.pone.0000607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Estrozi LF, Boehringer D, Shan S, Ban N, Schaffitzel C. Cryo-EM structure of the E. coli translating ribosome in complex with SRP and its receptor. Nat Struct Mol Biol. 2011;18:88–90. doi: 10.1038/nsmb.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang X, Kung S, Shan S. Demonstration of a two-step mechanism for assembly of the SRP-SRP receptor complex: implications for the catalytic role of SRP RNA. J Mol Biol. 2008;38:1:581–593. doi: 10.1016/j.jmb.2008.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Lam VQ, Mou Y, Kimura T, Chung J, Chandrasekar S, Winkler J, Mayo S, Shan S. Direct visualization reveals dynamics of a transient intermediate during protein assembly. Proc Natl Acad Sci USA. 2011;108:6450–6455. doi: 10.1073/pnas.1019051108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egea PF, Shan S, Napetschnig J, Savage DF, Walter P, Stroud RM. Substrate twinning activates the signal recognition particle and its receptor. Nature. 2004;427:215–221. doi: 10.1038/nature02250. [DOI] [PubMed] [Google Scholar]
- 16.Focia PJ, Shepotinovskaya IV, Seidler JA, Freymann DM. Heterodimeric GTPase core of the SRP targeting complex. Science. 2004;303:373–377. doi: 10.1126/science.1090827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shan S, Stroud R, Walter P. Mechanism of association and reciprocal activation of two GTPases. PLoS Biol. 2004;2:e320. doi: 10.1371/journal.pbio.0020320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, Schaffitzel C, Ban N, Shan S. Multiple conformational changes in a GTPase complex regulate protein targeting. Proc Natl Acad Sci USA. 2009;106:1754–1759. doi: 10.1073/pnas.0808573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen K, Zhang X, Shan S. Synergiestic action between the SRP RNA and translating ribosome allows efficient delivery of correct cargos during cotranslational protein targeting. RNA. 2011;17:892–902. doi: 10.1261/rna.2610411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang X, Rashid R, Wang K, Shan S. Sequential checkpoints govern fidelity during cotranslational protein targeting. Science. 2010;328:757–760. doi: 10.1126/science.1186743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lam VQ, Akopian D, Rome M, Shen Y, Henningsen D, Shan S. Lipid activation of the SRP receptor provides spatial coordination of protein targeting. J Cell Biol. 2010;190:623–635. doi: 10.1083/jcb.201004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shan S, Chandrasekar S, Walter P. Conformational changes in the GTPase modules of SRP and its receptor drive initiation of protein translocation. J Cell Biol. 2007;178:611–620. doi: 10.1083/jcb.200702018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halic M, Blau M, Becker T, Mielke T, Pool MR, Wild K, Sinning I, Beckmann R. Following the signal sequence from ribosomal tunnel exit to signal recognition particle. Nature. 2006;444:507–511. doi: 10.1038/nature05326. [DOI] [PubMed] [Google Scholar]
- 24.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
- 25.Attaide SF, Schmitz N, Shenk K, Ke A, Shan S, Doudna A, Ban N. The crystal structure of the signal recognition particle in complex with its receptor. Science. 2011;381:881–886. doi: 10.1126/science.1196473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaiser CA, Preuss D, Grisafi P, Botstein D. Many random sequences functionally replace the secretion signal sequence of yeast invertase. Science. 1987;235:312–317. doi: 10.1126/science.3541205. [DOI] [PubMed] [Google Scholar]
- 27.von Heijne G. Signal sequences: the limits of variation. J Mol Biol. 1985;184:99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]
- 28.Zheng N, Gierasch LM. Signal sequences: the same yet different. Cell. 1996;86:849–852. doi: 10.1016/s0092-8674(00)80159-2. [DOI] [PubMed] [Google Scholar]