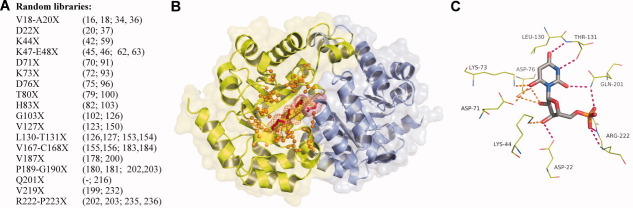
Figure 3.
A: List of the amino acid residue positions randomized in each library. In parenthesis, preceding the semicolon, are the corresponding residue positions in Methanothermobacter thermautotrophicus and following the semicolon, the positions in Saccharomyces cerevisiae. B: E. coli ODCase structure; Chain A is shown in green and Chain B is shown in blue (PDB code: 1EIX).17 The positions of randomized amino acid residues in and around one of the active sites are shown in orange. Completely conserved residues Lys44-Asp71-Lys73-Asp76 and their van der Waals radii are shown in red. C: Interaction of the barbituric acid inhibitor BMP (gray) with surrounding active site residues. Orange lines highlight interactions with the highly conserved network of charged residues. Magenta lines highlight the hydrogen bond network. Figures produced using PyMol.48