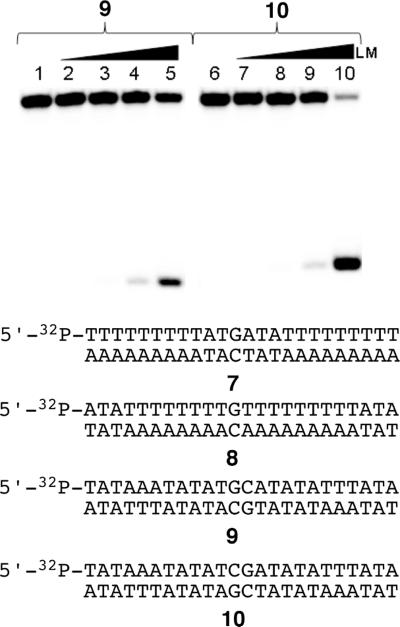
Figure 7.
Alkylation of palindromic DNA sequences by activated leinamycin. The palindromic duplexes 5'-32P-TAT AAA TAT ATG CAT ATA TTT ATA (9) and 5'-32P-TAT AAA TAT ATC GAT ATA TTT ATA (10) were incubated with leinamycin and 2-mercaptoethanol at 30 °C for 23 h in HEPES buffer (50 mM, pH 7) DETAPAC (5 mM), followed by Maxam-Gilbert workup and separation of the resulting DNA fragments on a 20% denaturing polyacrylamide gel. Reactions analyzed in lanes 1–5 contained duplex 9, while lanes 6–10 contained duplex 10. Reactions analyzed in lanes 1 and 6 contained DNA treated with 2-mercaptoethanol (5 mM, no leinamycin); lanes 2 and 7, contained DNA treated with leinamycin (10 μM) and 2-mercaptoethanol (5 μM); lanes 3 and 8, contained DNA treated with leinamycin (100 μM) and 2-mercaptoethanol (50 μM); lanes 4 and 9, contained DNA treated with leinamycin (1 mM) and 2-mercaptoethanol (0.5 mM); lanes 5 and 10 contained DNA treated with leinamycin (10 mM) and 2-mercaptoethanol (5 mM).