Figure 2.
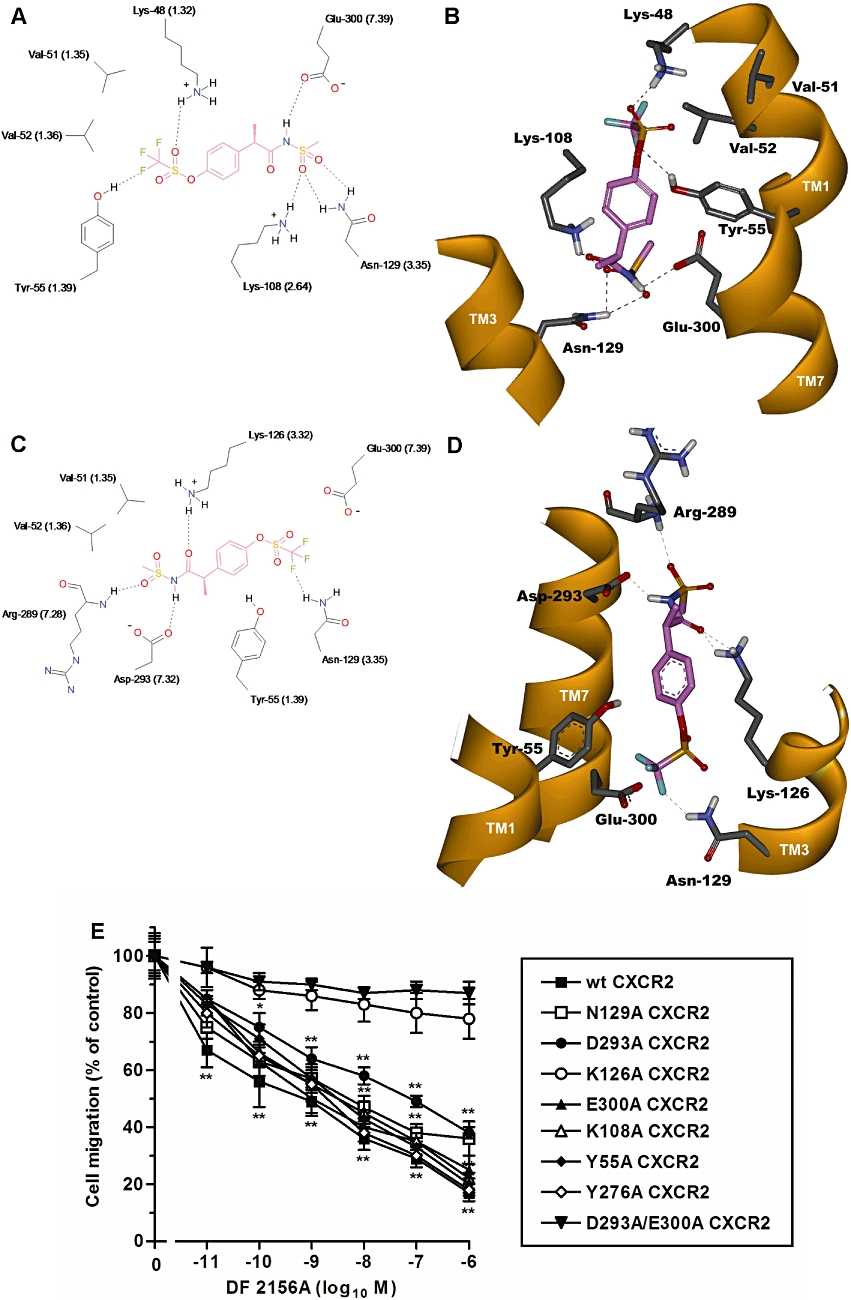
Molecular modelling of DF 2156A interaction with CXCR2. The two alternative binding modes DF 2156A in the CXCR2 allosteric pocket are shown in 2D (A, C) and 3D (B, D) representations viewed from within the plane of the membrane. 2D representation of DF 2156A (A, C) (pink) within the allosteric binding pocket of CXCR2. The most relevant amino acids and binding interactions in the two alternative orientations are depicted. Whereas in the first binding hypothesis (A) the key interactions of the CXCR1 binding mode (Glu291 TM7, Lys99 TM2 and Asn120 TM3) are conserved, in the alternative binding orientation (C), the acylmethanesulphonamide group establishes a strong network of polar interactions with Asp293 TM7 and Lys126 TM3, engaging also the backbone of Arg289 in the extracellular loop. Hydrogen bonds are represented as dashed lines. 3D representation of DF 2156A within the allosteric binding pocket of CXCR2 according to the alternative hypotheses (B, D). The amino acids involved in the hydrogen bond network engaged with the ligand (pink) are displayed, whereas the trans membrane domain is depicted as solid ribbon (orange). For a better comparison of CXCR1 and CXCR2, relative binding sites the Weinstein–Ballesteros nomenclature has been added in parenthses in the 2D representation. (E) Effect of DF 2156A on the IL-8-mediated migration of L1.2 transfectants expressing wild-type CXCR2 (wt CXCR2) or CXCR2 mutants. L1.2 transfectants were pre-incubated for 15 min at 37°C with vehicle or increasing concentrations of DF 2156A and then stimulated with 10 nM IL-8. Data are expressed as % of migration observed in the absence of DF 2156A (mean ± SD of three independent experiments). *P < 0.05 and **P < 0.01 versus cell migration in the absence of DF 2156A by Mann–Whitney U-test.
