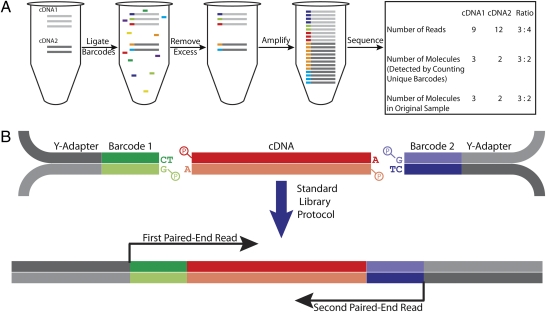
Fig. 1.
Our scheme of digital RNA-Seq. (A) General principle of digital RNA-Seq. Assume the original sample contains two cDNA sequences, one with three copies and another with two copies. An overwhelming number of unique barcode sequences are added to the sample in excess, and five are randomly ligated to the cDNA molecules. Ideally, each cDNA molecule in the sample receives a unique barcode sequence. After removing the excess barcodes, the barcoded cDNA molecules are amplified by PCR. Because of intrinsic noise and sequence-dependent bias, the barcoded cDNA molecules are amplified unevenly. Consequently, after the amplicons are sequenced, it appears that there are three copies of cDNA1 for every four copies of cDNA2 based on the relative number of reads for each sequence. However, the ratio in the original sample was 3:2, which is accurately reflected in the relative number of unique barcodes associated with each cDNA sequence. (B) In our implementation of A, we found it advantageous to randomly ligate both ends of each phosphorylated cDNA fragment to a barcoded phosphorylated Illumina Y-shaped adapter. Note that the single T and A overhangs present on the barcodes and cDNA, respectively, are to enhance ligation efficiency. After this step, the sample is amplified by PCR and prepared for sequencing using the standard Illumina library protocol. For each amplicon, both barcode sequences and both strands of the cDNA sequence are read using paired-end deep sequencing.