Fig. 2.
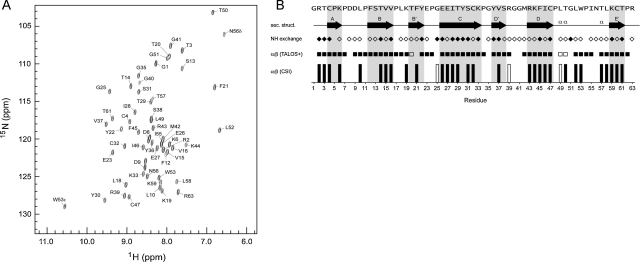
NMR analysis of β2GPI native DI. (A) The 2D 15N,1H-HSQC spectrum of 15N,13C-labelled β2GPI native DI. The sequence-specific backbone amide NH cross peak assignments obtained by 3D triple resonance methods are indicated. The dispersion of cross peaks is typical of a well-ordered globular protein domain. (B) Analysis of the β2GPI native DI NMR data with reference to the conformation in the X-ray structure of intact β2GPI. We plot two indicators on a per-residue basis of the local backbone torsion angles based upon the backbone atom chemical shift assignments for β2GPI DI: the chemical shift index (CSI; bottom) and the secondary structure prediction of the program TALOS+. In each case the prediction is either for extended strand (filled symbol) or α-helical (empty symbol) conformation. Also indicated is the qualitative assessment of the rate of backbone amide NH/solvent exchange due to H-bonding based upon the presence or absence of an exchange NH/solvent water cross peak in a 3D 15N-separated 1H-NOESY spectrum of β2GPI DI. The overall pattern of these parameters is consistent with the β-strand-rich secondary structure of β2GPI DI (top) as seen in the crystal structure (PDB code 1QUB32).